2O9Z
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the Apo form | | Descriptor: | PHOSPHATE ION, Tryptophan halogenase | | Authors: | Bitto, E, Bingman, C.A, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
4GNU
 
 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase GES-5 | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2012-08-17 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
4KGC
 
 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | Descriptor: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | Authors: | Adhireksan, Z, Davey, C.A. | | Deposit date: | 2013-04-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
6WF0
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H3 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6WEZ
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14-D93N Mutant Bound to the Influenza A H3 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
1MMO
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL NON-HAEM IRON HYDROXYLASE THAT CATALYSES THE BIOLOGICAL OXIDATION OF METHANE | | Descriptor: | ACETIC ACID, FE (III) ION, METHANE MONOOXYGENASE HYDROLASE (ALPHA CHAIN), ... | | Authors: | Rosenzweig, A.C, Frederick, C.A, Lippard, S.J, Nordlund, P. | | Deposit date: | 1994-02-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane.
Nature, 366, 1993
|
|
4GOG
 
 | | Crystal structure of the GES-1 imipenem acyl-enzyme complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase GES-1, IODIDE ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Munoz, J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
2MZZ
 
 | | NMR structure of APOBEC3G NTD variant, sNTD | | Descriptor: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | Authors: | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | Deposit date: | 2015-02-28 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4J8U
 
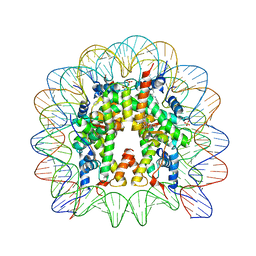 | |
6DGX
 
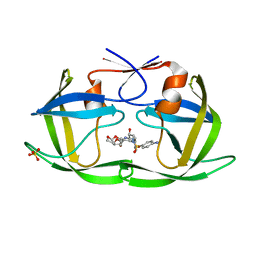 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH6
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DID
 
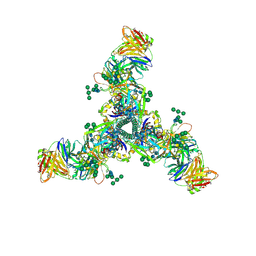 | | HIV Env BG505 SOSIP with polyclonal Fabs from immunized rabbit #3417 post-boost#1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Turner, H.L, Cottrell, C.A, Oyen, D, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
2G08
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product-transition complex analog with Aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
1MND
 
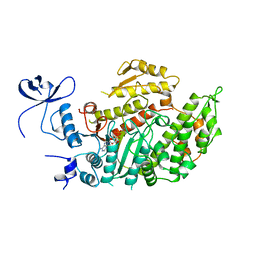 | |
3LZV
 
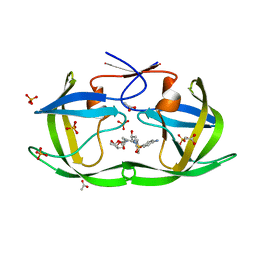 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Kolli, M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
6DET
 
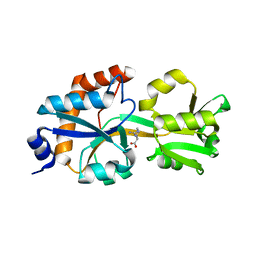 | |
6WF1
 
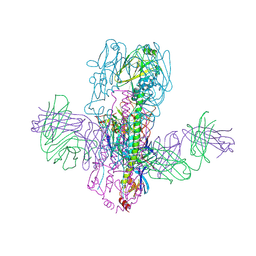 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H10 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
3M5L
 
 | | Crystal structure of HCV NS3/4A protease in complex with ITMN-191 | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3/4A, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4F75
 
 | |
6WQY
 
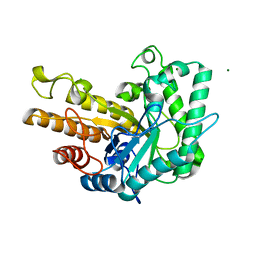 | | GH5-4 broad specificity endoglucanase from Bacteroides salanitronis | | Descriptor: | CHLORIDE ION, Cellulase, MAGNESIUM ION | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
4L5F
 
 | |
6CZR
 
 | | The structure of amicetin bound to the 70S ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Eiler, D.R, Steitz, T.A, Looper, R.E, Serrano, C.M, Kannareddy, H.R, Koch, M.R, Barrows, L.R, Testa, C.A, Sebahar, P.R. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Unifying the Aminohexopyranose- and Peptidyl-Nucleoside Antibiotics: Implications for Antibiotic Design
Angew.Chem.Int.Ed.Engl., 132, 2020
|
|
3MGP
 
 | | Binding of Cobalt ions to the Nucleosome Core Particle | | Descriptor: | CHLORIDE ION, COBALT (II) ION, DNA (147-MER), ... | | Authors: | Mohideen, K, Muhammad, R, Davey, C.A. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
