6DH8
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DIS
 
 | | Crystal structure of HCV NS3/4A protease in complex with P4-1 (AJ-71) | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5, 16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6- yl]carbamate, GLYCEROL, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6WBB
 
 | | Structure of Mouse Importin alpha - MLH1-E475A NLS peptide complex | | Descriptor: | DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | De Barros, A.C, Da Silva, T.D, Oliveira, H.C, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.663 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
6WBA
 
 | | Structure of Mouse Importin alpha MLH1- R470A NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | de Barros, A.C, da Silva, T.D, Oliveira, H.C, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
2OAZ
 
 | | Human Methionine Aminopeptidase-2 Complexed with SB-587094 | | Descriptor: | COBALT (II) ION, N-(2-ISOPROPYLPHENYL)-3-[(2-THIENYLMETHYL)THIO]-1H-1,2,4-TRIAZOL-5-AMINE, human Methionine Amino Peptidase 2 | | Authors: | Marino Jr, J.P, Fisher, P.W, Hofmann, G.A, Kirkpatrick, R, Janson, C.A, Johnson, R.K, Ma, C, Mattern, M, Meek, T.D, Ryan, D, Schulz, C, Smith, W.W, Tew, D.G, Tomazek Jr, T.A, Veber, D.F, Xiong, W.C, Yamamoto, Y, Yamashita, K, Yang, G, Thompson, S.K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore.
J.Med.Chem., 50, 2007
|
|
6WBC
 
 | | Structure of Mouse Importin alpha- MLH1-R472K NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | de Barros, A.C, da Silva, T.D, Oliveira, H.C, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
3N4U
 
 | |
2NYI
 
 | | Crystal Structure of an Unknown Protein from Galdieria sulphuraria | | Descriptor: | unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tandem ACT domain-containing protein ACTP from Galdieria sulphuraria.
Proteins, 80, 2012
|
|
2FZ9
 
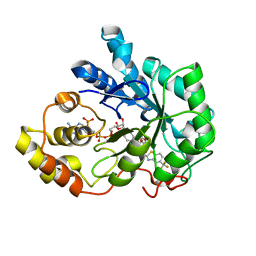 | | Human Aldose Reductase complexed with inhibitor zopolrestat after six days soaking. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Gerlach, C, Sotriffer, C.A, Heine, A, Klebe, G. | | Deposit date: | 2006-02-09 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions.
J.Mol.Biol., 363, 2006
|
|
2G09
 
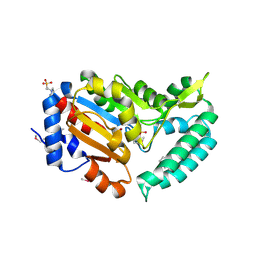 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product complex | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G13
 
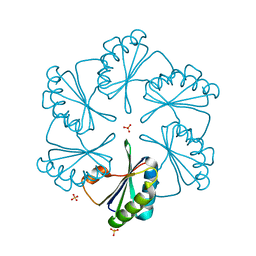 | | CsoS1A with sulfate ion | | Descriptor: | Major carboxysome shell protein 1A, SULFATE ION | | Authors: | Tsai, Y, Sawaya, M.R, Cannon, G.C, Williams, E.B, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2006-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome.
Plos Biol., 5, 2007
|
|
1PJU
 
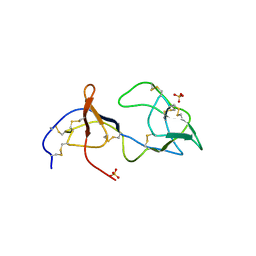 | | Unbound form of Tomato Inhibitor-II | | Descriptor: | SULFATE ION, Wound-induced proteinase inhibitor II | | Authors: | Barrette-Ng, I.H, Ng, K.K.-S, Cherney, M.M, Pearce, G, Ghani, U, Ryan, C.A, James, M.N.G. | | Deposit date: | 2003-06-03 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unbound form of tomato inhibitor-II reveals interdomain flexibility and conformational variability in the reactive site loops
J.Biol.Chem., 278, 2003
|
|
2NZD
 
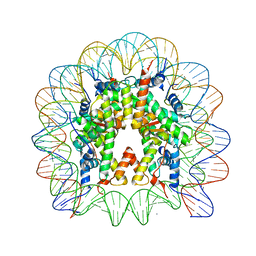 | | Nucleosome core particle containing 145 bp of DNA | | Descriptor: | DNA (145-MER), Histone H2B, Histone H3, ... | | Authors: | Ong, M.S, Richmond, T.J, Davey, C.A. | | Deposit date: | 2006-11-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA stretching and extreme kinking in the nucleosome core
J.Mol.Biol., 368, 2007
|
|
1OJI
 
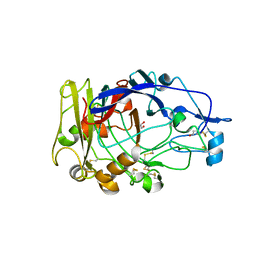 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
4L9Z
 
 | | Crystal Structure of Rhodobacter sphaeroides malyl-CoA lyase in complex with magnesium, oxalate, and CoA | | Descriptor: | COENZYME A, MAGNESIUM ION, Malyl-CoA lyase, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
6VLU
 
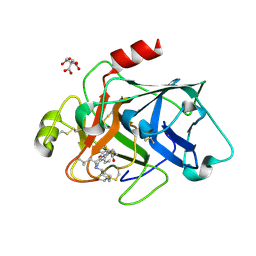 | | Factor XIa in complex with compound 7 | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, N-cyclohexyl-D-leucyl-N-[(1-aminoisoquinolin-6-yl)methyl]-4,4-difluoro-L-prolinamide | | Authors: | Lesburg, C.A, Fradera, X. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-06 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery.
J.Chem.Inf.Model., 60, 2020
|
|
3NIA
 
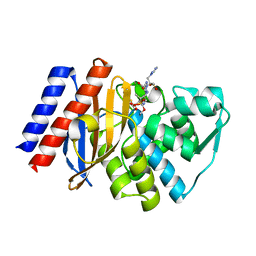 | | GES-2 carbapenemase tazobactam complex | | Descriptor: | (3S)-3-(dihydroxy-lambda~4~-sulfanyl)-4-(1H-1,2,3-triazol-1-yl)-D-valine, Beta-lactamase GES-2, TAZOBACTAM INTERMEDIATE | | Authors: | Frase, H, Smith, C.A, Toth, M, Vakulenko, S.B. | | Deposit date: | 2010-06-15 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of the GES-2 beta-Lactamse by Tazobactam: Detection of Reaction
Intermediates by UV and Mass Spectrometry and X-Ray Crystallography
To be Published
|
|
1OIR
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 1-(DIMETHYLAMINO)-3-(4-{{4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)PYRIMIDIN-2-YL]AMINO}PHENOXY)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4JO8
 
 | | Crystal structure of the activating Ly49H receptor in complex with m157 (G1F strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor 8, M157 | | Authors: | Berry, R, Ng, N, Saunders, P.M, Vivian, J.P, Lin, J, Deuss, F.A, Corbett, A.J, Forbes, C.A, Widjaja, J.M, Sullivan, L.C, McAlister, A.D, Perugini, M.A, Call, M.J, Scalzo, A.A, Degli-Esposti, M.A, Coudert, J.D, Beddoe, T, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting of a natural killer cell receptor family by a viral immunoevasin
Nat.Immunol., 14, 2013
|
|
2FZD
 
 | | Human aldose reductase complexed with tolrestat at 1.08 A resolution. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Gerlach, C, Sotriffer, C.A, Heine, A, Klebe, G. | | Deposit date: | 2006-02-09 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions.
J.Mol.Biol., 363, 2006
|
|
1OR7
 
 | | Crystal Structure of Escherichia coli sigmaE with the Cytoplasmic Domain of its Anti-sigma RseA | | Descriptor: | RNA polymerase sigma-E factor, Sigma-E factor negative regulatory protein | | Authors: | Campbell, E.A, Tupy, J.L, Gruber, T.M, Wang, S, Sharp, M.M, Gross, C.A, Darst, S.A. | | Deposit date: | 2003-03-12 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA.
Mol.Cell, 11, 2003
|
|
2GCB
 
 | | G51S/S52T double mutant of L. casei FPGS | | Descriptor: | Folylpolyglutamate synthase | | Authors: | Smith, C.A, Cross, J.A, Bognar, A.L, Sun, X. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutation of Gly51 to serine in the P-loop of Lactobacillus casei folylpolyglutamate synthetase abolishes activity by altering the conformation of two adjacent loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3N4V
 
 | |
1OYV
 
 | | Crystal structure of tomato inhibitor-II in a ternary complex with subtilisin Carlsberg | | Descriptor: | CALCIUM ION, Subtilisin Carlsberg, Wound-induced proteinase inhibitor-II | | Authors: | Barrette-Ng, I.H, Ng, K.K, Cherney, M.M, Pearce, G, Ryan, C.A, James, M.N. | | Deposit date: | 2003-04-07 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg
J.Biol.Chem., 278, 2003
|
|
6C74
 
 | |
