2K95
 
 | | Solution structure of the wild-type P2B-P3 pseudoknot of human telomerase RNA | | Descriptor: | Telomerase RNA P2b-P3 pseudoknot | | Authors: | Kim, N.-K, Zhang, Q, Zhou, J, Theimer, C.A, Peterson, R.D, Feigon, J. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA.
J.Mol.Biol., 384, 2008
|
|
1XLE
 
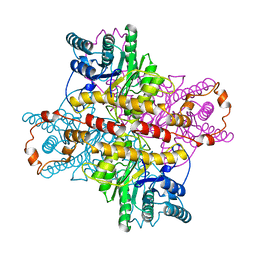 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
6Q2N
 
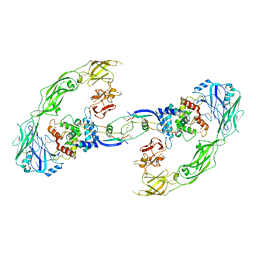 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6N6U
 
 | |
8B8V
 
 | |
2KO8
 
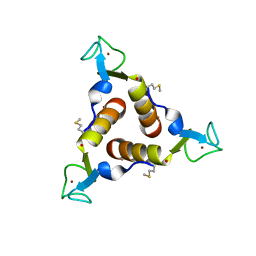 | | The Structure of Anti-TRAP | | Descriptor: | Tryptophan RNA-binding attenuator protein inhibitory protein, ZINC ION | | Authors: | McElroy, C.A, Gollnick, P, Foster, M.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-09-22 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B. subtilis Anti-TRAP trimer and its Interaction with TRAP
To be Published
|
|
2KRF
 
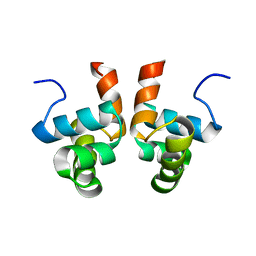 | | NMR solution structure of the DNA binding domain of Competence protein A | | Descriptor: | Transcriptional regulatory protein comA | | Authors: | Hobbs, C.A, Bobay, B.G, Thompson, R.J, Perego, M, Cavanagh, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and DNA-binding model of the DNA-binding domain of competence protein A.
J.Mol.Biol., 398, 2010
|
|
6NLU
 
 | |
2I3F
 
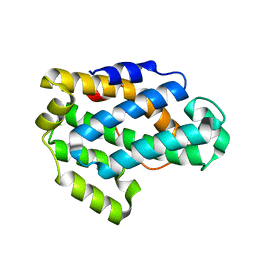 | | Crystal Structure of a Glycolipid transfer-like protein from Galdieria sulphuraria | | Descriptor: | glycolipid transfer-like protein | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of a Glycolipid transfer-like protein from Galdieria sulphuraria
To be Published
|
|
2I5T
 
 | | Crystal Structure of hypothetical protein LOC79017 from Homo sapiens | | Descriptor: | Protein C7orf24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
5U2P
 
 | | The crystal structure of Tp0737 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Norgard, M.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Functional clues from the crystal structure of an orphan periplasmic ligand-binding protein from Treponema pallidum.
Protein Sci., 26, 2017
|
|
3SBC
 
 | | Crystal structure of Saccharomyces cerevisiae TSA1C47S mutant protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Peroxiredoxin TSA1 | | Authors: | Tairum Jr, C.A, Horta, B.B, Netto, L.E.S, Oliveira, M.A. | | Deposit date: | 2011-06-03 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disulfide biochemistry in 2-cys peroxiredoxin: requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin.
J.Mol.Biol., 424, 2012
|
|
2I1P
 
 | | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 2 | | Authors: | Wolf, C.A, Dancea, F, Shi, M, Bade-Noskova, V, Rueterjans, H, Kerjaschki, D, Luecke, C. | | Deposit date: | 2006-08-14 | | Release date: | 2007-02-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin.
J.Biomol.Nmr, 37, 2007
|
|
2I0D
 
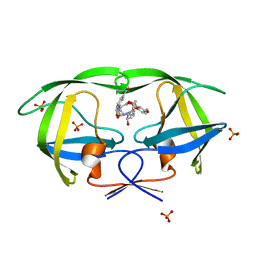 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2I3C
 
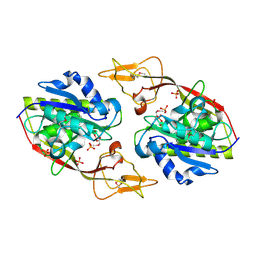 | | Crystal Structure of an Aspartoacylase from Homo Sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I5S
 
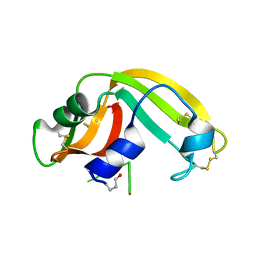 | | Crystal structure of onconase with bound nucleic acid | | Descriptor: | 5'-D(*A*(DU)P*GP*A)-3', P-30 protein | | Authors: | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
4F74
 
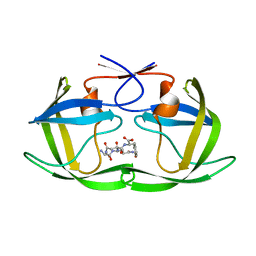 | |
4EYM
 
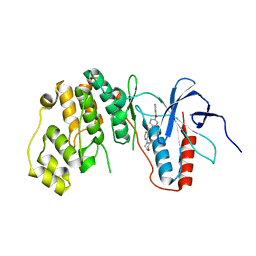 | | MAPK13 complex with inhibitor | | Descriptor: | 2-(morpholin-4-yl)-N-[4-(pyridin-4-yloxy)phenyl]pyridine-4-carboxamide, Mitogen-activated protein kinase 13 | | Authors: | Miller, C.A, Brett, T.J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | IL-13-induced airway mucus production is attenuated by MAPK13 inhibition.
J.Clin.Invest., 122, 2012
|
|
1D17
 
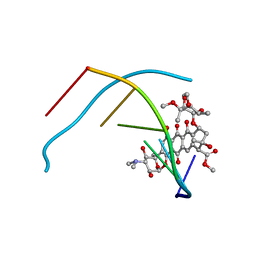 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
4FE3
 
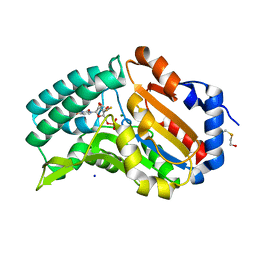 | |
2BDU
 
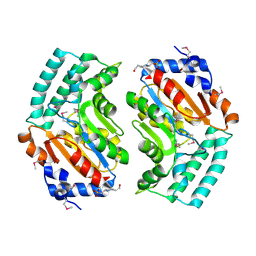 | | X-Ray Structure of a Cytosolic 5'-Nucleotidase III from Mus Musculus MM.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2BE4
 
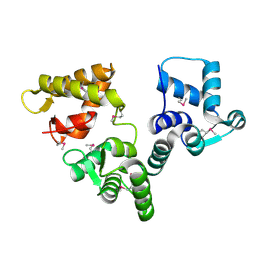 | | X-RAY STRUCTURE AN EF-HAND PROTEIN FROM DANIO RERIO Dr.36843 | | Descriptor: | hypothetical protein LOC449832 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-21 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Danio rerio secretagogin: A hexa-EF-hand calcium sensor.
Proteins, 76, 2009
|
|
5UUO
 
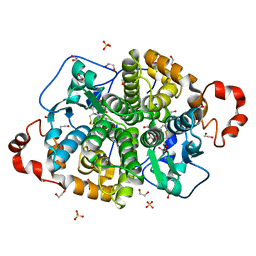 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase-like protein, ... | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
6OAL
 
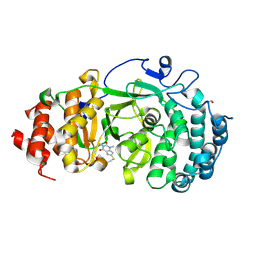 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Ahmed, Z, Warden, S, Tainer, J.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6QVJ
 
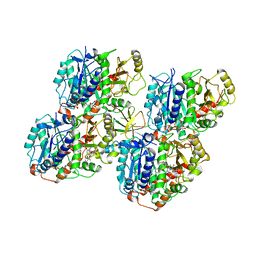 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
