6O84
 
 | | Cryo-EM structure of OTOP3 from xenopus tropicalis | | Descriptor: | LOC100127796 protein,LOC100127796 protein,OTOP3,LOC100127796 protein,LOC100127796 protein | | Authors: | Chen, Q.F, Bai, X.C, Jiang, Y.X. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional characterization of an otopetrin family proton channel.
Elife, 8, 2019
|
|
6XPL
 
 | | CutR Screw, form 2 with 33.8 angstrom pitch | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6YY4
 
 | | Parallel 17-mer DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*AP*AP*GP*GP*GP*TP*GP*GP*GP*A)-3') | | Authors: | Srb, P, Curtis, C, Veverka, V. | | Deposit date: | 2020-05-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Overlapping but distinct: a new model for G-quadruplex biochemical specificity.
Nucleic Acids Res., 49, 2021
|
|
8CIU
 
 | | The FERM domain of human moesin mutant H288A | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
4WR5
 
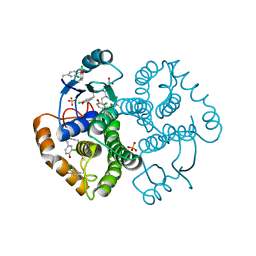 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
1C1U
 
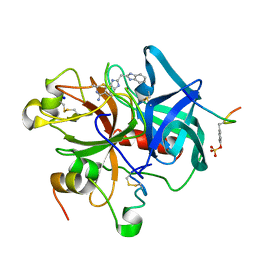 | |
4KEF
 
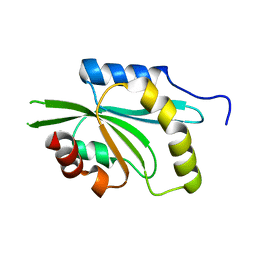 | |
1RY4
 
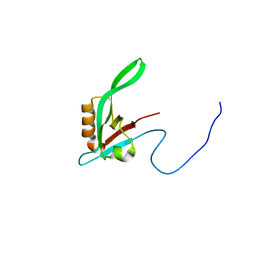 | |
8OZC
 
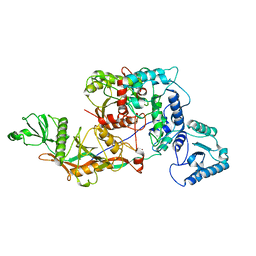 | | cryoEM structure of SPARTA complex heterodimer apo | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Ekundayo, B, Ni, D.C, Lu, X.H, Stahlberg, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
2REJ
 
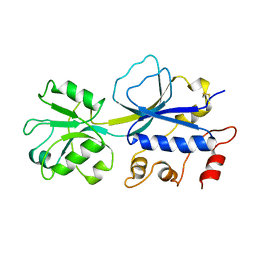 | | ABC-transporter choline binding protein in unliganded semi-closed conformation | | Descriptor: | PUTATIVE GLYCINE BETAINE ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2RNK
 
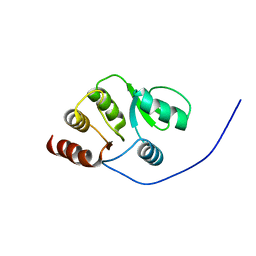 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1S4T
 
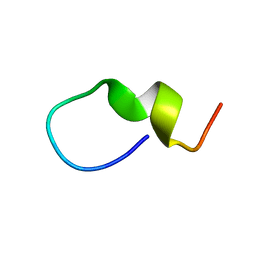 | | Solution structure of synthetic 21mer peptide spanning region 135-155 (in human numbering) of sheep prion protein | | Descriptor: | Major prion protein | | Authors: | Kozin, S.A, Lepage, C, Hui Bon Hoa, G, Rabesona, H, Mazur, A.K, Blond, A, Cheminant, M, Haertle, T, Debey, P, Rebuffat, S. | | Deposit date: | 2004-01-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific recognition between surface loop 2 (132-143) and helix 1 (144-154)
within sheep prion protein from in vitro studies of synthetic peptides
To be Published
|
|
3LXU
 
 | |
4WP7
 
 | | Structure of human ALDH1A1 with inhibitor CM026 | | Descriptor: | 8-{[4-(furan-2-ylcarbonyl)piperazin-1-yl]methyl}-1,3-dimethyl-7-(3-methylbutyl)-3,7-dihydro-1H-purine-2,6-dione, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of Two Distinct Structural Classes of Selective Aldehyde Dehydrogenase 1A1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
1IIJ
 
 | |
4X47
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Anhydrosialidase, PHOSPHATE ION | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4X4L
 
 | | Structure of human ALDH1A1 with inhibitor CM037 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-12-03 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of Two Distinct Structural Classes of Selective Aldehyde Dehydrogenase 1A1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
3CW5
 
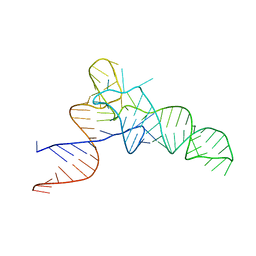 | | E. coli Initiator tRNA | | Descriptor: | Initiator tRNA | | Authors: | Barraud, P, Schmitt, E, Mechulam, Y, Dardel, F, Tisne, C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis.
Nucleic Acids Res., 36, 2008
|
|
1CCW
 
 | |
1S5Q
 
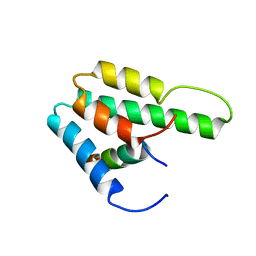 | | Solution Structure of Mad1 SID-mSin3A PAH2 Complex | | Descriptor: | MAD protein, Sin3a protein | | Authors: | Swanson, K.A, Knoepfler, P.S, Huang, K, Kang, R.S, Cowley, S.M, Laherty, C.D, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2004-01-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4WO0
 
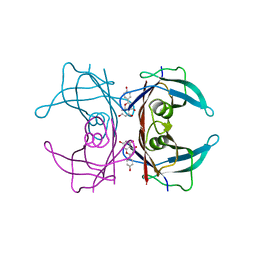 | | Crystal structure of transthyretin in complex with apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Folli, C, Berni, R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5HQ6
 
 | | Crystal structure of fragment bound with Brd4 | | Descriptor: | (2R)-2,6-dimethyl-2H-1,4-benzoxazin-3(4H)-one, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Xu, Y.C. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of fragment bound with Brd4
to be published
|
|
2RM6
 
 | | Glutathione peroxidase-type tryparedoxin peroxidase, reduced form | | Descriptor: | Glutathione peroxidase-like protein | | Authors: | Melchers, J, Feher, K, Diechtierow, M, Krauth-Siegel, L, Muhle-Goll, C. | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for a distinct catalytic mechanism in Trypanosoma brucei tryparedoxin peroxidase
J.Biol.Chem., 283, 2008
|
|
5HFG
 
 | | Cytosolic disulfide reductase DsbM from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein, cytosolic disulfide reductase DsbM | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Crystal structures of the disulfide reductase DsbM from Pseudomonas aeruginosa
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4WUT
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
