4AAA
 
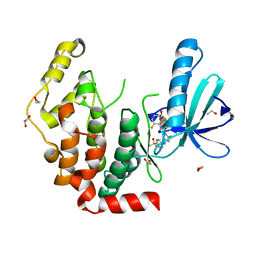 | | Crystal structure of the human CDKL2 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CYCLIN-DEPENDENT KINASE-LIKE 2 | | Authors: | Canning, P, Vollmar, M, Cooper, C.D.O, Mahajan, P, Daga, N, Berridge, G, Burgess-Brown, N, Muniz, J.R.C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
4ABN
 
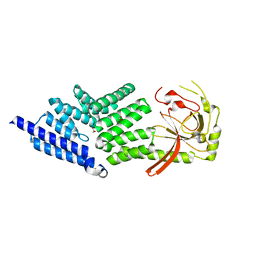 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BKN
 
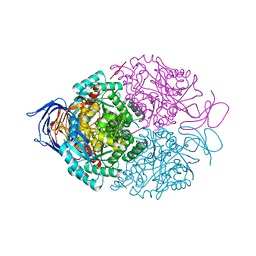 | | Human Dihydropyrimidinase-related protein 3 (DPYSL3) | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 3 | | Authors: | Mathea, S, Elkins, J.M, Alegre-Abarrategui, J, Shrestha, L, Burgess-Brown, N, Puranik, S, Coutandin, D, Bradley, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Dpysl3
To be Published
|
|
4AGU
 
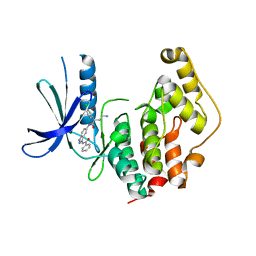 | | CRYSTAL STRUCTURE OF THE HUMAN CDKL1 KINASE DOMAIN | | Descriptor: | CYCLIN-DEPENDENT KINASE-LIKE 1, N-(5-{[(2S)-4-amino-2-(3-chlorophenyl)butanoyl]amino}-1H-indazol-3-yl)benzamide | | Authors: | Canning, P, Sharpe, T.D, Allerston, C, Savitsky, P, Pike, A.C.W, Muniz, J.R.C, Chaikuad, A, Kuo, K, Burgess-Brown, N, Ayinampudi, V, Zhang, Y, Thangaratnarajah, C, Ugochukwu, E, Vollmar, M, Krojer, T, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Knapp, S, Bullock, A. | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
4BBM
 
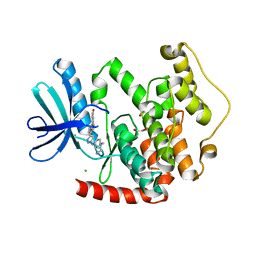 | | CRYSTAL STRUCTURE OF THE HUMAN CDKL2 KINASE DOMAIN WITH BOUND TCS 2312 | | Descriptor: | 1,2-ETHANEDIOL, 4'-[5-[[3-[(CYCLOPROPYLAMINO)METHYL]PHENYL]AMINO]-1H-PYRAZOL-3-YL]-[1,1'-BIPHENYL]-2,4-DIOL, CHLORIDE ION, ... | | Authors: | Canning, P, Elkins, J.M, Cooper, C.D.O, Mahajan, P, Daga, N, Berridge, G, Burgess-Brown, N, Muniz, J.R.C, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A. | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
4BZY
 
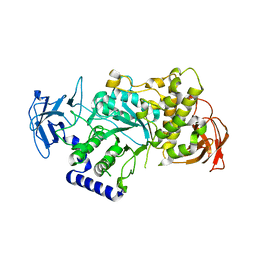 | | Crystal structure of human glycogen branching enzyme (GBE1) | | Descriptor: | 1,4-ALPHA-GLUCAN-BRANCHING ENZYME | | Authors: | Froese, D.S, Krojer, T, Goubin, S, Strain-Damerell, C, Mahajan, P, von Delft, F, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W.W. | | Deposit date: | 2013-07-30 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Glycogen Branching Enzyme Deficiency and Pharmacologic Rescue by Rational Peptide Design.
Hum.Mol.Genet., 24, 2015
|
|
3ZP5
 
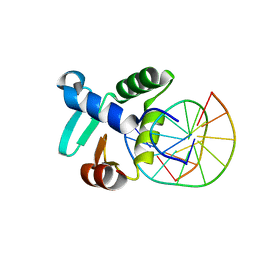 | | Crystal structure of the DNA binding ETS domain of the human protein FEV in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', PROTEIN FEV | | Authors: | Newman, J.A, Cooper, C.D.O, Krojer, T, Shrestha, L, Allerston, C.K, Vollmar, M, Arrowsmith, C.H, Burgess-Brown, N, Edwards, A, von Delft, F, Gileadi, O. | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4CVH
 
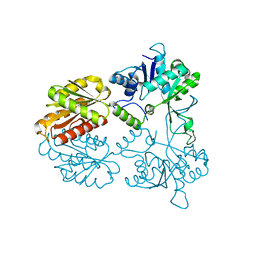 | | Crystal structure of human isoprenoid synthase domain-containing protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPRENOID SYNTHASE DOMAIN-CONTAINING PROTEIN, ... | | Authors: | Kopec, J, Froese, D.S, Krojer, T, Newman, J, Kiyani, W, Goubin, S, Strain-Damerell, C, Vollmar, M, von Delft, F, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Lefeber, D.J, Yue, W.W. | | Deposit date: | 2014-03-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Human Ispd is a Cytidyltransferase Required for Dystroglycan O-Mannosylation.
Chem.Biol., 22, 2015
|
|
5G6V
 
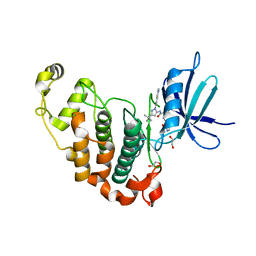 | | Crystal structure of the PCTAIRE1 kinase in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, CYCLIN-DEPENDENT KINASE 16 | | Authors: | Dixon-Clarke, S.E, Galan Bartual, S, Elkins, J, Savitsky, P, Kopec, J, Mackenzie, A, Tallant, C, Heroven, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
5HES
 
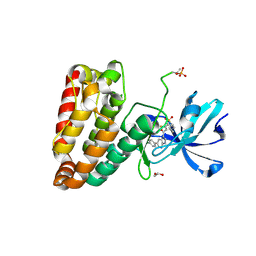 | | Human leucine zipper- and sterile alpha motif-containing kinase (ZAK, MLT, HCCS-4, MRK, AZK, MLTK) in complex with vemurafenib | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase MLT, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | Authors: | Mathea, S, Salah, E, Abdul Azeez, K.R, Tallant, C, Szklarz, M, Chaikuad, A, Shrestha, B, Sorrell, F.J, Elkins, J.M, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of the Human Protein Kinase ZAK in Complex with Vemurafenib.
Acs Chem.Biol., 11, 2016
|
|
5FDP
 
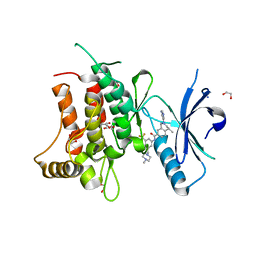 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
5FDX
 
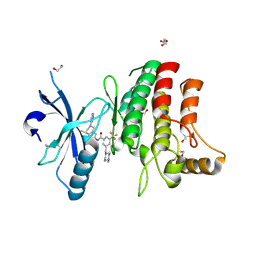 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
5FII
 
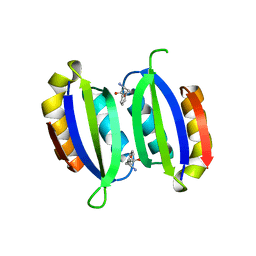 | | Structure of a human aspartate kinase, chorismate mutase and TyrA domain. | | Descriptor: | PHENYLALANINE, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Patel, D, Kopec, J, Shrestha, L, Fitzpatrick, F, Pinkas, D, Chaikuad, A, Dixon-Clarke, S, McCorvie, T.J, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Ligand-Dependent Dimerization of Phenylalanine Hydroxylase Regulatory Domain.
Sci.Rep., 6, 2016
|
|
4AD9
 
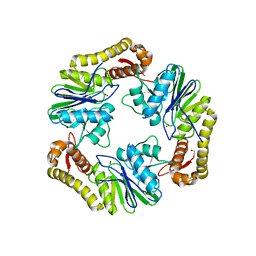 | | Crystal structure of human LACTB2. | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE-LIKE PROTEIN 2, ZINC ION | | Authors: | Allerston, C.K, Krojer, T, Shrestha, B, Burgess Brown, N, Chalk, R, Elkins, J.M, Filippakopoulos, P, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Lactb2, a Metallo-Beta-Lactamase Protein, as a Human Mitochondrial Endoribonuclease
Nucleic Acids Res., 44, 2016
|
|
5O5E
 
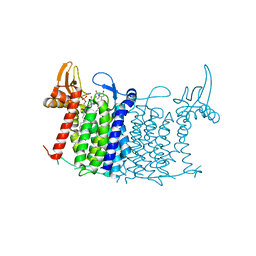 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with tunicamycin | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5Q1W
 
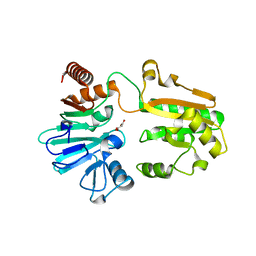 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000546a | | Descriptor: | 1~{H}-benzimidazol-2-ylcyanamide, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QJ4
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1827602749 | | Descriptor: | (2S,3S)-3-methyl-N-(1,2,3-thiadiazol-5-yl)tetrahydrofuran-2-carboxamide (non-preferred name), 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJL
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z56983806 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-N-(3-methylphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJB
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1787627869 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-methyl-N-{[(3R)-oxolan-3-yl]methyl}pyrimidin-4-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJO
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z57292369 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QK8
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1271660837 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z94597856 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJV
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z281802060 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-N-methylpyrimidin-2-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJM
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z328695024 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QK2
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z54628578 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
