7N1M
 
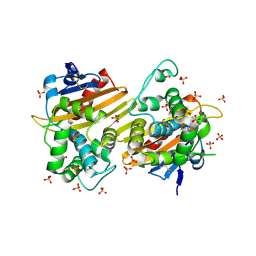 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Orthorhombic Crystal Form | | 分子名称: | Beta-lactamase OXA-935, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.B, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-27 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
4BJH
 
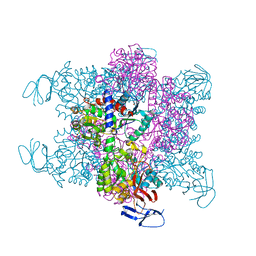 | | Crystal Structure of the Aquifex Reactor Complex Formed by Dihydroorotase (H180A, H232A) with Dihydroorotate and Aspartate Transcarbamoylase with N-(phosphonacetyl)-L-aspartate (PALA) | | 分子名称: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, ASPARTATE CARBAMOYLTRANSFERASE, ... | | 著者 | Edwards, B.F.P, Martin, P.D, Grimley, E, Vaishnav, A, Fernando, R, Brunzelle, J.S, Cordes, M, Evans, H.G, Evans, D.R. | | 登録日 | 2013-04-18 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Mononuclear Metal Center of Type-I Dihydroorotase from Aquifex Aeolicus.
Bmc Biochem., 14, 2013
|
|
3M5A
 
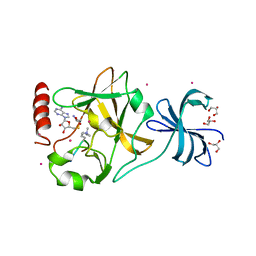 | | SET7/9 Y245A in complex with TAF10-K189me3 peptide and AdoHcy | | 分子名称: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | 著者 | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | 登録日 | 2010-03-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M59
 
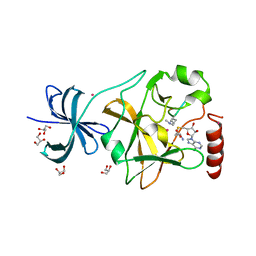 | | SET7/9 Y245A in complex with TAF10-K189me2 peptide and AdoHcy | | 分子名称: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | 著者 | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | 登録日 | 2010-03-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M55
 
 | | SET7/9 Y305F in complex with TAF10-K189me1 peptide and AdoHcy | | 分子名称: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | 著者 | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | 登録日 | 2010-03-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M53
 
 | | SET7/9 in complex with TAF10 peptide and AdoHcy | | 分子名称: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | 著者 | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | 登録日 | 2010-03-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M58
 
 | | SET7/9 Y245A in complex with TAF10-K189me1 peptide and AdoHcy | | 分子名称: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | 登録日 | 2010-03-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3DGK
 
 | |
3DFC
 
 | |
3DFI
 
 | |
3DFK
 
 | |
3CHU
 
 | | Crystal Structure of Di-iron Aurf | | 分子名称: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | 著者 | Zhang, H, Brunzelle, J.S, Nair, S.K. | | 登録日 | 2008-03-10 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CHT
 
 | |
7JIJ
 
 | | ATP-bound AMP-activated protein kinase | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | 登録日 | 2020-07-23 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | 分子名称: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-10-04 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | 分子名称: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-10-04 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
6MON
 
 | | Crystal structure of human SMYD2 in complex with Nle-peptide inhibitor | | 分子名称: | GLYCEROL, LYS-LEU-NLE-SER-LYS-ARG-GLY, N-lysine methyltransferase SMYD2, ... | | 著者 | Spellmon, N, Cornett, E, Brunzelle, J, Rothbart, S, Yang, Z. | | 登録日 | 2018-10-04 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.711 Å) | | 主引用文献 | A functional proteomics platform to reveal the sequence determinants of lysine methyltransferase substrate selectivity.
Sci Adv, 4, 2018
|
|
5EQV
 
 | | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-MALATE, FE (III) ION, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-13 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site.
To Be Published
|
|
4RRU
 
 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | 分子名称: | CALCIUM ION, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-06 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RS9
 
 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | 分子名称: | Protein TIFY 7, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
6WVN
 
 | | Crystal Structure of Nsp16-Nsp10 from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-Adenosylmethionine. | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ADENINE, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-06 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WQ3
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-adenosyl-L-homocysteine. | | 分子名称: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-28 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WRZ
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 with 7-methyl-GpppA and S-adenosyl-L-homocysteine in the Active Site and Sulfates in the mRNA Binding Groove. | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-30 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
4PXQ
 
 | | Crystal structure of D-glucuronyl C5-epimerase in complex with heparin hexasaccharide | | 分子名称: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, D-glucuronyl C5 epimerase B | | 著者 | Ke, J, Qin, Y, Gu, X, Tan, J, Brunzelle, J.S, Xu, H.E, Ding, K. | | 登録日 | 2014-03-24 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
5JHP
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | 分子名称: | Protein TPR1, The rice D53 EAR peptide (794-808) | | 著者 | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | 登録日 | 2016-04-21 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
