1GX5
 
 | |
1GX6
 
 | |
7Q5G
 
 | | LAN-DAP5 DERIVATIVE OF LANREOTIDE: L-DIAMINO PROPIONIC ACID IN POSITION 5 IN PLACE OF L-LYSINE | | Descriptor: | ETHANOL, LAN-DAP5 DERIVATIVE OF LANREOTIDE | | Authors: | Bressanelli, S, Le Du, M.H, Gobeaux, F, Legrand, P, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1CSJ
 
 | | CRYSTAL STRUCTURE OF THE RNA-DEPENDENT RNA POLYMERASE OF HEPATITIS C VIRUS | | Descriptor: | HEPATITIS C VIRUS RNA POLYMERASE (NS5B) | | Authors: | Bressanelli, S, Tomei, L, Roussel, A, Incitti, I, Vitale, R.L, Mathieu, M, De Francesco, R, Rey, F.A. | | Deposit date: | 1999-08-18 | | Release date: | 1999-11-08 | | Last modified: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1URZ
 
 | |
7Q5A
 
 | | Lanreotide nanotube | | Descriptor: | Lanreotide | | Authors: | Pieri, L, Wang, F, Arteni, A.A, Bressanelli, S, Egelman, E.H, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8PWH
 
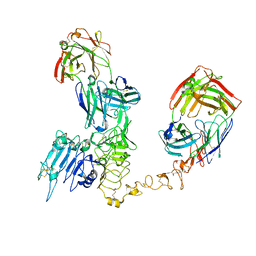 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
8Q6J
 
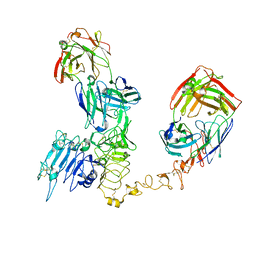 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
6YPT
 
 | |
3I5K
 
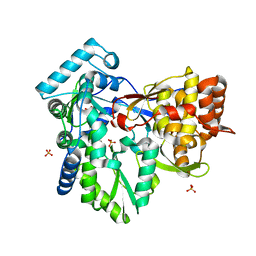 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
4TT1
 
 | |
4TT0
 
 | |
5LW5
 
 | |
5LWA
 
 | |
2J6E
 
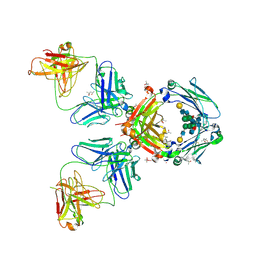 | | Crystal Structure of an Autoimmune Complex between a Human IgM Rheumatoid Factor and IgG1 Fc reveals a Novel Fc Epitope and Evidence for Affinity Maturation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CACODYLATE ION, ... | | Authors: | Duquerroy, S, Stura, E.A, Bressanelli, S, Browne, H, Beale, D, Hamon, M, Casali, P, Vaney, M.C, Rey, F.A, Sutton, B.J, Taussig, M.J. | | Deposit date: | 2006-09-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a human autoimmune complex between IgM rheumatoid factor RF61 and IgG1 Fc reveals a novel epitope and evidence for affinity maturation.
J.Mol.Biol., 368, 2007
|
|
1WCE
 
 | | Crystal structure of the T13 IBDV viral particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1WCD
 
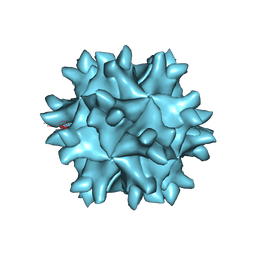 | | Crystal structure of IBDV T1 virus-like particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
5MDM
 
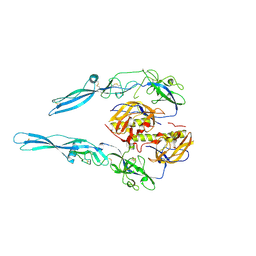 | | Structural intermediates in the fusion associated transition of vesiculovirus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Baquero, E, Albertini, A.A, Gaudin, Y, Bressanelli, S. | | Deposit date: | 2016-11-11 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural intermediates in the fusion-associated transition of vesiculovirus glycoprotein.
EMBO J., 36, 2017
|
|
5I2S
 
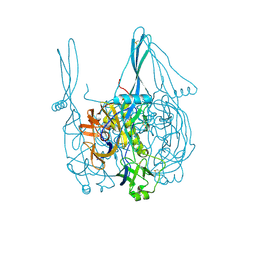 | |
5I2M
 
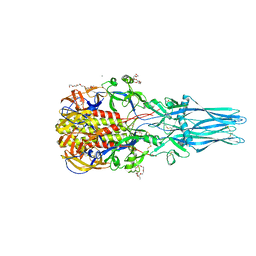 | |
4A5U
 
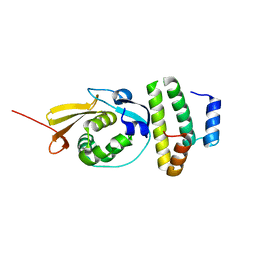 | |
4ADP
 
 | | HCV-J6 NS5B POLYMERASE V405I MUTANT | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Scrima, N, Bressanelli, S. | | Deposit date: | 2012-01-02 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Two Crucial Early Steps in RNA Synthesis by the Hepatitis C Virus Polymerase Involve a Dual Role of Residue 405.
J.Virol., 86, 2012
|
|
4AEP
 
 | |
4AEX
 
 | |
2XYM
 
 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
