5IVO
 
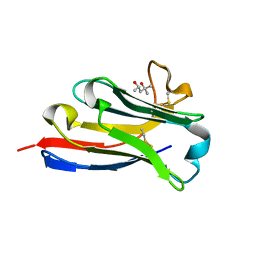 | | BC2 nanobody | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BC2-nanobody | | Authors: | Braun, M.B, Stehle, T. | | Deposit date: | 2016-03-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptides in headlock - a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy.
Sci Rep, 6, 2016
|
|
8B5A
 
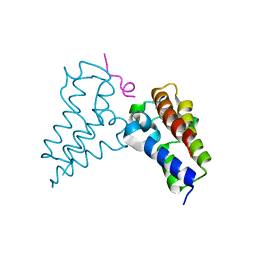 | | Human BRD3 bromodomain 2 in complex with a H4 peptide containing ApmTri (H4K20ApmTri) | | Descriptor: | Bromodomain-containing protein 3, H4K20ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8B5B
 
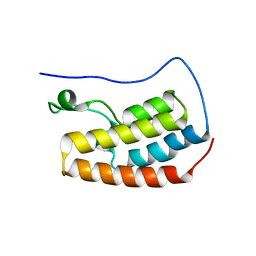 | | Human BRD4 bromdomain 1 in complex with a H4 peptide containing acetyl lysine and ApmTri (H4K5acK8ApmTri) | | Descriptor: | Bromodomain-containing protein 4, DI(HYDROXYETHYL)ETHER, H4K5acK8ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8B5C
 
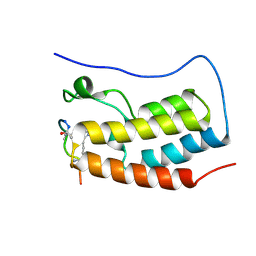 | | Human BRD4 bromdomain 1 in complex with a H4 peptide containing ApmTri (H4K5/8ApmTri) | | Descriptor: | Bromodomain-containing protein 4, H4K5/8ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6QJU
 
 | |
8PKU
 
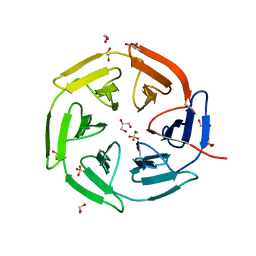 | | Kelch domain of KEAP1 in complex with ortho-dimethylbenzene linked cyclic peptide 3 (ortho-WRCDEETGEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8PKW
 
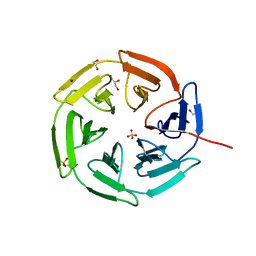 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 5 (ortho-WRCDPETaEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8PKX
 
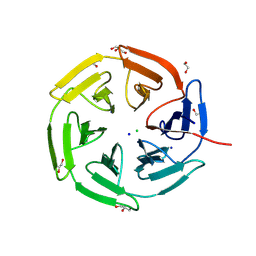 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 11 (ortho-WRCNPETaEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8PKV
 
 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 4 (ortho-WRCDEETGEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
5IVN
 
 | | BC2 nanobody in complex with the BC2 peptide tag | | Descriptor: | BC2-nanobody, Cadherin derived peptide | | Authors: | Braun, M.B, Stehle, T. | | Deposit date: | 2016-03-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Peptides in headlock - a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy.
Sci Rep, 6, 2016
|
|
8UVL
 
 | | Crystal structure of selective IRE1a inhibitor 29 at the enzyme active site | | Descriptor: | 1,2-ETHANEDIOL, 1-phenyl-N-(2,3,6-trifluoro-4-{[(3M)-3-(2-{[(3R,5R)-5-fluoropiperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}phenyl)methanesulfonamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Kiefer, J.R, Wallweber, H.A, Braun, M.-G, Wei, W, Jiang, F, Wang, W, Rudolph, J, Ashkenazi, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Potent, Selective, and Orally Available IRE1 alpha Inhibitors Demonstrating Comparable PD Modulation to IRE1 Knockdown in a Multiple Myeloma Model.
J.Med.Chem., 67, 2024
|
|
8EXL
 
 | | Crystal structure of PI3K-alpha in complex with taselisib | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXO
 
 | | Crystal structure of PI3K-alpha in complex with compound 19 | | Descriptor: | 1-{(4S,11aM)-2-[(4R)-2-oxo-4-(propan-2-yl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-prolinamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXU
 
 | | Crystal structure of PI3K-alpha in complex with compound 30 | | Descriptor: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXV
 
 | | Crystal structure of PI3K-alpha in complex with compound 32 | | Descriptor: | N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8A92
 
 | | p53-Y220C Core Domain in Complex with a Bromo-trifluoro-pyrazole-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromanyl-5-(trifluoromethyl)-1H-pyrazol-3-amine, Cellular tumor antigen p53, ... | | Authors: | Stahlecker, J, Braun, M.B, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C.
Rsc Med Chem, 13, 2022
|
|
1RM8
 
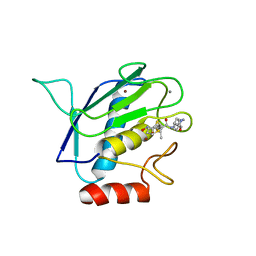 | | Crystal structure of the catalytic domain of MMP-16/MT3-MMP: Characterization of MT-MMP specific features | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, Matrix metalloproteinase-16, ... | | Authors: | Lang, R, Braun, M, Sounni, N.E, Noel, A, Frankenne, F, Foidart, J.-M, Bode, W, Maskos, K. | | Deposit date: | 2003-11-27 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic domain of MMP-16/MT3-MMP: characterization of MT-MMP specific features.
J.Mol.Biol., 336, 2004
|
|
1KFX
 
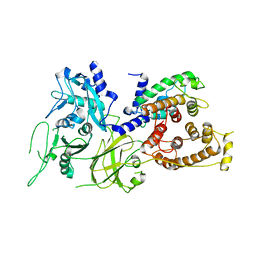 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KFU
 
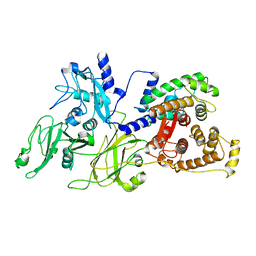 | | Crystal Structure of Human m-Calpain Form II | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1JK3
 
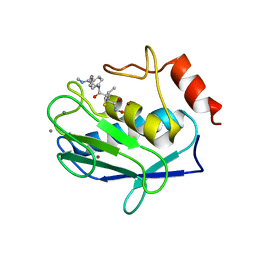 | | Crystal structure of human MMP-12 (Macrophage Elastase) at true atomic resolution | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, MACROPHAGE METALLOELASTASE, ... | | Authors: | Lang, R, Kocourek, A, Braun, M, Tschesche, H, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Substrate specificity determinants of human macrophage elastase (MMP-12) based on the 1.1 A crystal structure.
J.Mol.Biol., 312, 2001
|
|
3J9I
 
 | | Thermoplasma acidophilum 20S proteasome | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Li, X, Mooney, P, Zheng, S, Booth, C, Braunfeld, M.B, Gubbens, S, Agard, D.A, Cheng, Y. | | Deposit date: | 2015-02-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Nat.Methods, 10, 2013
|
|
4UAQ
 
 | | Crystal structure of the accessory translocation ATPase, SecA2, from Mycobacterium tuberculosis | | Descriptor: | Protein translocase subunit SecA 2 | | Authors: | Swanson-Smith, S, Ioerger, T.R, Rigel, N.W, Miller, B.K, Braunstein, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2.
J.Bacteriol., 198, 2015
|
|
1NL3
 
 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS in APO FORM | | Descriptor: | PREPROTEIN TRANSLOCASE SECA 1 SUBUNIT | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NKT
 
 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEX WITH ADPBS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase secA 1 subunit | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-03 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
