3HW2
 
 | | Crystal structure of the SifA-SKIP(PH) complex | | Descriptor: | Pleckstrin homology domain-containing family M member 2, Protein sifA | | Authors: | Diacovich, L, Dumont, A, Lafitte, D, Soprano, E, Guilhon, A.-A, Bignon, C, Gorvel, J.-P, Bourne, Y, Meresse, S. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Interaction between the SifA virulence factor and its host target skip is essential for salmonella pathogenesis
J.Biol.Chem., 284, 2009
|
|
7QZO
 
 | | Crystal structure of GacS D1 domain | | Descriptor: | CADMIUM ION, GLYCEROL, Histidine kinase | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
7QZ2
 
 | | Crystal structure of GacS D1 domain in complex with BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CADMIUM ION, Histidine kinase, ... | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
8QTL
 
 | | Aplysia californica acetylcholine-binding protein in complex with Spiroimine (-)-4 S | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Soluble acetylcholine receptor, ... | | Authors: | Sulzenbacher, G, Bourne, Y, Marchot, P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cyclic Imine Core Common to the Marine Macrocyclic Toxins Is Sufficient to Dictate Nicotinic Acetylcholine Receptor Antagonism.
Mar Drugs, 22, 2024
|
|
8QX2
 
 | |
5NN8
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
8Q1M
 
 | | Aplysia californica acetylcholine-binding protein in complex with Spiroimine (+)-4 R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Soluble acetylcholine receptor, ... | | Authors: | Sulzenbacher, G, Bourne, Y, Marchot, P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cyclic Imine Core Common to the Marine Macrocyclic Toxins Is Sufficient to Dictate Nicotinic Acetylcholine Receptor Antagonism.
Mar Drugs, 22, 2024
|
|
5NN6
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-hydroxyethyl-1-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
5NN4
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-acetyl-cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
5NN5
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
4AEH
 
 | | Crystal structure of the anti-AaHI Fab9C2 antibody | | Descriptor: | FAB ANTIBODY, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
4AEI
 
 | | Crystal structure of the AaHII-Fab4C1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALPHA-MAMMAL TOXIN AAH2, CHLORIDE ION, ... | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | Descriptor: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4URI
 
 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
3BE8
 
 | | Crystal structure of the synaptic protein neuroligin 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-01-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and Its beta-Neurexin Complex: Determinants for Folding and Cell Adhesion
Neuron, 56, 2007
|
|
7Z8N
 
 | | GacS histidine kinase from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, Histidine kinase, R-1,2-PROPANEDIOL | | Authors: | Fadel, F, Bassim, V, Francis, V.I, Porter, S.L, Botzanowski, T, Legrand, P, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
6HZZ
 
 | | Structure of human D-glucuronyl C5 epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1ODU
 
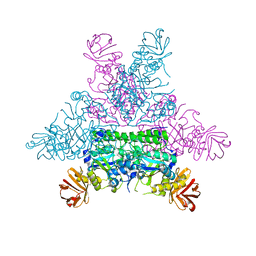 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH FUCOSE | | Descriptor: | PUTATIVE ALPHA-L-FUCOSIDASE, beta-L-fucopyranose | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
1AVA
 
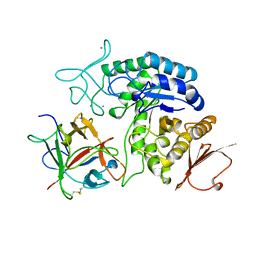 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
6I01
 
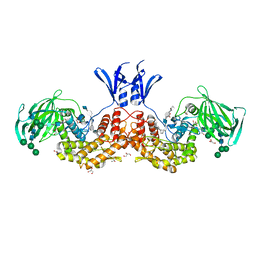 | | Structure of human D-glucuronyl C5 epimerase in complex with substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I02
 
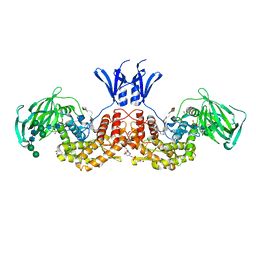 | | Structure of human D-glucuronyl C5 epimerase in complex with product | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4AWN
 
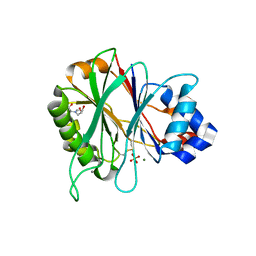 | | Structure of recombinant human DNase I (rhDNaseI) in complex with Magnesium and Phosphate. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parsiegla, G, Noguere, C, Santell, L, Lazarus, R.A, Bourne, Y. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Human DNase I Bound to Magnesium and Phosphate Ions Points to a Catalytic Mechanism Common to Members of the DNase I-Like Superfamily.
Biochemistry, 51, 2012
|
|
4QDN
 
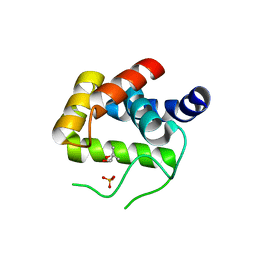 | | Crystal Structure of the endo-beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Flagellar protein FlgJ [peptidoglycan hydrolase], PHOSPHATE ION | | Authors: | Lipski, A, Nurizzo, D, Bourne, Y, Vincent, F. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of the beta-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family.
Glycobiology, 25, 2015
|
|
2BYO
 
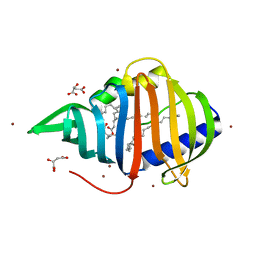 | | Crystal structure of Mycobacterium tuberculosis lipoprotein LppX (Rv2945c) | | Descriptor: | ACETATE ION, ALPHA-LINOLENIC ACID, D-MALATE, ... | | Authors: | Sulzenbacher, G, Canaan, S, Roig-Zamboni, V, Maurin, D, Gicquel, B, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lppx is a Lipoprotein Required for the Translocation of Phthiocerol Dimycocerosates to the Surface of Mycobacterium Tuberculosis.
Embo J., 25, 2006
|
|
1Q40
 
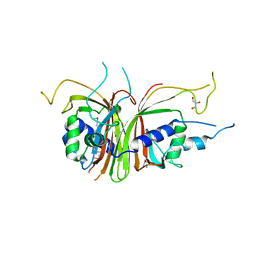 | | Crystal structure of the C. albicans Mtr2-Mex67 M domain complex | | Descriptor: | GLYCEROL, MRNA TRANSPORT REGULATOR Mtr2, mRNA export factor MEX67 | | Authors: | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | Deposit date: | 2003-08-01 | | Release date: | 2003-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
