1MPP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
1KS9
 
 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
1N0W
 
 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
1KA0
 
 | |
1JP4
 
 | | Crystal Structure of an Enzyme Displaying both Inositol-Polyphosphate 1-Phosphatase and 3'-Phosphoadenosine-5'-Phosphate Phosphatase Activities | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Patel, S, Yenush, L, Rodriguez, P.L, Serrano, R, Blundell, T.L. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of an enzyme displaying both inositol-polyphosphate-1-phosphatase and 3'-phosphoadenosine-5'-phosphate phosphatase activities: a novel target of lithium therapy.
J.Mol.Biol., 315, 2002
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1Z56
 
 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
1YJQ
 
 | | Crystal structure of ketopantoate reductase in complex with NADP+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-dehydropantoate 2-reductase, ACETATE ION, ... | | Authors: | Lobley, C.M.C, Ciulli, A, Whitney, H.M, Williams, G, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of Escherichia coli ketopantoate reductase with NADP+ bound.
Biochemistry, 44, 2005
|
|
1YON
 
 | | Escherichia coli ketopantoate reductase in complex with 2-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2-dehydropantoate 2-reductase, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5R)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Ciulli, A, Lobley, C.M.C, Tuck, K.L, Williams, G, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2005-01-28 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | pH-tuneable binding of 2'-phospho-ADP-ribose to ketopantoate reductase: a structural and calorimetric study.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1XOD
 
 | | Crystal structure of X. tropicalis Spred1 EVH-1 domain | | Descriptor: | GLYCEROL, Spred1 | | Authors: | Harmer, N.J, Sivak, J.M, Amaya, E, Blundell, T.L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | 1.15A Crystal structure of the X. tropicalis Spred1 EVH1 domain suggests a fourth distinct peptide-binding mechanism within the EVH1 family
Febs Lett., 579, 2005
|
|
2ASU
 
 | |
1YYH
 
 | | Crystal structure of the human Notch 1 ankyrin domain | | Descriptor: | Notch 1, ankyrin domain | | Authors: | Ehebauer, M.T, Chirgadze, D.Y, Hayward, P, Martinez-Arias, A, Blundell, T.L. | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | High-resolution crystal structure of the human Notch 1 ankyrin domain
Biochem.J., 392, 2005
|
|
2BB2
 
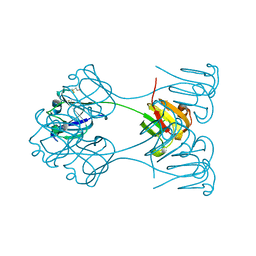 | | X-RAY ANALYSIS OF BETA B2-CRYSTALLIN AND EVOLUTION OF OLIGOMERIC LENS PROTEINS | | Descriptor: | BETA B2-CRYSTALLIN, BETA-MERCAPTOETHANOL | | Authors: | Bax, B, Lapatto, R, Nalini, V, Driessen, H, Lindley, P.F, Mahadevan, D, Blundell, T.L, Slingsby, C. | | Deposit date: | 1992-09-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray analysis of beta B2-crystallin and evolution of oligomeric lens proteins.
Nature, 347, 1990
|
|
1PDA
 
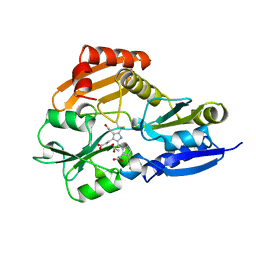 | | STRUCTURE OF PORPHOBILINOGEN DEAMINASE REVEALS A FLEXIBLE MULTIDOMAIN POLYMERASE WITH A SINGLE CATALYTIC SITE | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, PORPHOBILINOGEN DEAMINASE | | Authors: | Louie, G.V, Brownlie, P.D, Lambert, R, Cooper, J.B, Blundell, T.L, Wood, S.P, Warren, M.J, Woodcock, S.C, Jordan, P.M. | | Deposit date: | 1992-11-17 | | Release date: | 1993-10-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Nature, 359, 1992
|
|
1QIB
 
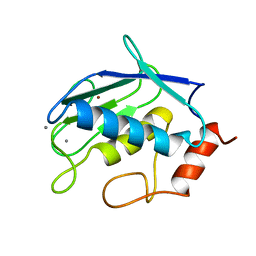 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
1QGX
 
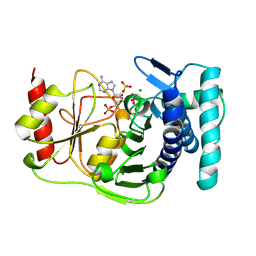 | | X-RAY STRUCTURE OF YEAST HAL2P | | Descriptor: | 3',5'-ADENOSINE BISPHOSPHATASE, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Albert, A, Yenush, L, Gil-Mascarell, M.R, Rodriguez, P.L, Patel, J, Martinez-Ripoll, M, Blundell, T.L, Serrano, R. | | Deposit date: | 1999-05-10 | | Release date: | 2000-01-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity.
J.Mol.Biol., 295, 2000
|
|
1QIA
 
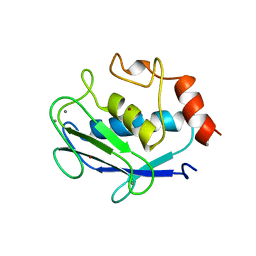 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1QIC
 
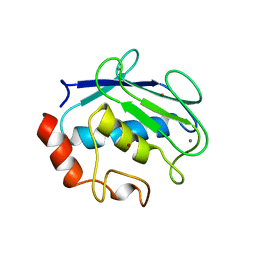 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1PWA
 
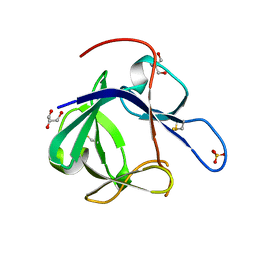 | | Crystal structure of Fibroblast Growth Factor 19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fibroblast growth factor-19, GLYCEROL, ... | | Authors: | Harmer, N.J, Pellegrini, L, Chirgadze, D, Fernandez-Recio, J, Blundell, T.L. | | Deposit date: | 2003-07-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity.
Biochemistry, 43, 2004
|
|
5CHP
 
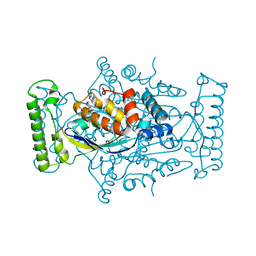 | | T. maritima ThyX in complex with TyC5-03 | | Descriptor: | (2S)-2-[(3-hydroxy-5-oxo-4,5-dihydro-1,2,4-triazin-6-yl)sulfanyl]propanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Surade, S, Luciani, R, Saxena, P, Santucci, M, Ferrari, S, Venturelli, A, Marverti, G, Ponterini, G, Abell, C.A, Blundell, T.L, Costi, M.P. | | Deposit date: | 2015-07-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of inhibitors targeting substrate-binding site in Mycobacterium tuberculosis FAD-dependent Thymidylate synthase (ThyX) through Virtual Screening: A New study de-fining the binding mechanism of Inhibitors
To Be Published
|
|
1RQF
 
 | | Structure of CK2 beta subunit crystallized in the presence of a p21WAF1 peptide | | Descriptor: | Casein kinase II beta chain, Disordered segment of Cyclin-dependent kinase inhibitor 1, UNKNOWN LIGAND, ... | | Authors: | Bertrand, L, Sayed, M.F, Pei, X.-Y, Parisini, E, Dhanaraj, V, Bolanos-Garcia, V.M, Allende, J.E, Blundell, T.L. | | Deposit date: | 2003-12-05 | | Release date: | 2004-10-05 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the regulatory subunit of CK2 in the presence of a p21WAF1 peptide demonstrates flexibility of the acidic loop.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3HWO
 
 | |
3IMG
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.8 Ang resolution in a ternary complex with fragment compounds 5-methoxyindole and 1-benzofuran-2-carboxylic acid | | Descriptor: | 1-benzofuran-2-carboxylic acid, 5-methoxy-1H-indole, ETHANOL, ... | | Authors: | Ciulli, A, Hung, A.W, Silvestre, H.L, Wen, S, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUB
 
 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-Methoxy-N-(5-methylpyridin-2-ylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-N-[(5-methylpyridin-2-yl)sulfonyl]-1H-indole-2-carboxamide, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUE
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(5-methoxy-2-(5-Methylpyridin-2-ylsulfonylcarbamoyl)-1H-indol-1-yl) acetic acid | | Descriptor: | (5-methoxy-2-{[(5-methylpyridin-2-yl)sulfonyl]carbamoyl}-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Wen, S, Hung, A.W, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
