5FOS
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH OLIGOMERISATION PEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, PHOSPHATE ION | | Authors: | Sharpe, T, Moschetti, T, Fischer, G, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5FOT
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHTU TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTU PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.189 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOU
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHPA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHPA PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOW
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOX
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHAA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHAA PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5IOY
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(cyclopentylmethyl)pyrrolidine-1-carboxamide at 1.77A resolution | | Descriptor: | N-(cyclopentylmethyl)pyrrolidine-1-carboxamide, SULFATE ION, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5IPA
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with (E)-3-(furan-3-yl)-1-(pyrrolidin-1-yl)prop-2-en-1-one at 1.78A resolution | | Descriptor: | (2E)-3-(furan-3-yl)-1-(pyrrolidin-1-yl)prop-2-en-1-one, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5IOZ
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(cyclopentylmethyl)cyclopentanecarboxamide at 2.02A resolution | | Descriptor: | N-(cyclopentylmethyl)cyclopentanecarboxamide, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5IP6
 
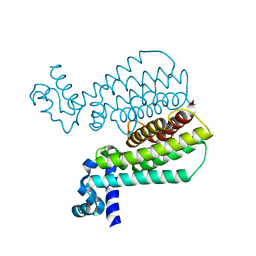 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-((tetrahydrofuran-3-yl)methyl)pyrrolidine-1-carboxamide at 1.93A resolution | | Descriptor: | N-{[(3S)-oxolan-3-yl]methyl}pyrrolidine-1-carboxamide, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
4CMS
 
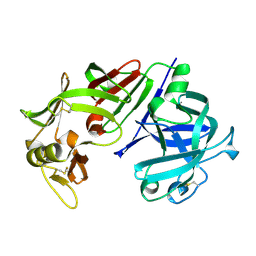 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES IV. STRUCTURE AND REFINEMENT AT 2.2 ANGSTROMS RESOLUTION OF BOVINE CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Khan, G, Tickle, I.J, Blundell, T.L, Safro, M, Andreeva, N, Zdanov, A. | | Deposit date: | 1991-11-01 | | Release date: | 1991-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin.
J.Mol.Biol., 221, 1991
|
|
4KIJ
 
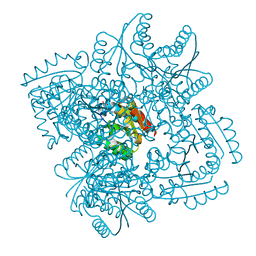 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinase dehydratase from Mycobacterium tuberculosis - compound 35c [3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid] | | Descriptor: | 3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KI7
 
 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinase from Mycobacterium tuberculosis - compound 41c [3-hydroxy-5-(3-nitrophenoxy)benzoic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 3-hydroxy-5-(3-nitrophenoxy)benzoic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-01 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KLX
 
 | |
4KIU
 
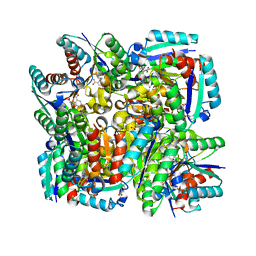 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49d [5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KG0
 
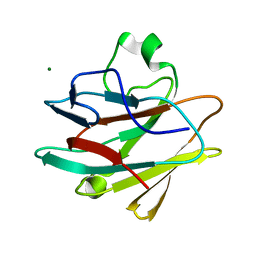 | | Crystal structure of the drosophila melanogaster neuralized-nhr1 domain | | Descriptor: | MAGNESIUM ION, Protein neuralized | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2013-04-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure, biochemical and biophysical characterisation of NHR1 domain of E3 Ubiquitin ligase neutralized
Advances in Enzyme Research, 1, 2013
|
|
4KM2
 
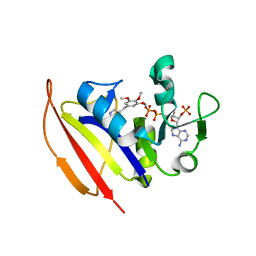 | |
4KM0
 
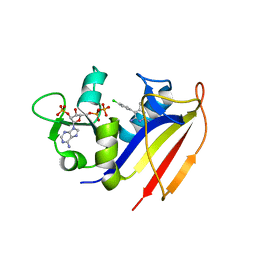 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with pyrimethamine | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KL9
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in the space group C2 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KIW
 
 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49e [5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
1CZI
 
 | | CHYMOSIN COMPLEX WITH THE INHIBITOR CP-113972 | | Descriptor: | CHYMOSIN, CP-113972 (NORSTATINE-S-METHYL CYSTEINE-IODO-PHENYLALANINE-PROLINE) | | Authors: | Groves, M.R, Dhanaraj, V, Pitts, J.E, Badasso, M, Hoover, D, Nugent, P, Blundell, T.L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 2.3 A resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active site beta-hairpin flap is rearranged when compared with the native crystal structure.
Protein Eng., 11, 1998
|
|
1EPO
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
1PQF
 
 | | Glycine 24 to Serine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYQ
 
 | | Unprocessed Aspartate Decarboxylase Mutant, with Alanine inserted at position 24 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PPY
 
 | | Native precursor of pyruvoyl dependent Aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase precursor, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
