5FOV
 
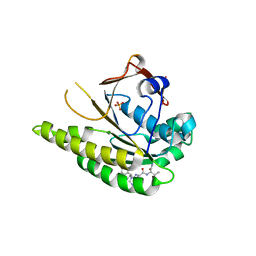 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHTG TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, FHTG PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOW
 
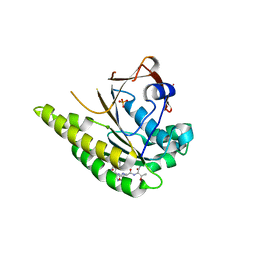 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5FPK
 
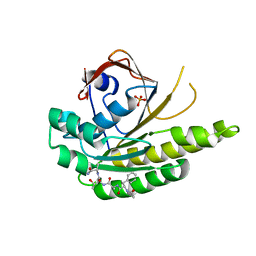 | | MONOMERIC RADA IN COMPLEX WITH FATA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTG PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
2ASU
 
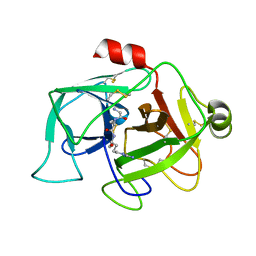 | |
5L8V
 
 | | Apo-structure of humanised RadA-mutant humRadA4 | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB4
 
 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
1YJQ
 
 | | Crystal structure of ketopantoate reductase in complex with NADP+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-dehydropantoate 2-reductase, ACETATE ION, ... | | Authors: | Lobley, C.M.C, Ciulli, A, Whitney, H.M, Williams, G, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of Escherichia coli ketopantoate reductase with NADP+ bound.
Biochemistry, 44, 2005
|
|
2BB2
 
 | | X-RAY ANALYSIS OF BETA B2-CRYSTALLIN AND EVOLUTION OF OLIGOMERIC LENS PROTEINS | | Descriptor: | BETA B2-CRYSTALLIN, BETA-MERCAPTOETHANOL | | Authors: | Bax, B, Lapatto, R, Nalini, V, Driessen, H, Lindley, P.F, Mahadevan, D, Blundell, T.L, Slingsby, C. | | Deposit date: | 1992-09-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray analysis of beta B2-crystallin and evolution of oligomeric lens proteins.
Nature, 347, 1990
|
|
3IUB
 
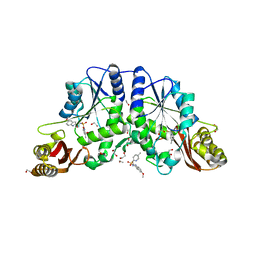 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-Methoxy-N-(5-methylpyridin-2-ylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-N-[(5-methylpyridin-2-yl)sulfonyl]-1H-indole-2-carboxamide, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUE
 
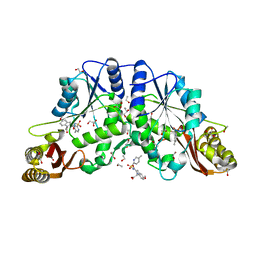 | | Crystal structure of pantothenate synthetase in complex with 2-(5-methoxy-2-(5-Methylpyridin-2-ylsulfonylcarbamoyl)-1H-indol-1-yl) acetic acid | | Descriptor: | (5-methoxy-2-{[(5-methylpyridin-2-yl)sulfonyl]carbamoyl}-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Wen, S, Hung, A.W, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5K4Z
 
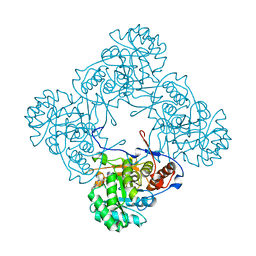 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5K4X
 
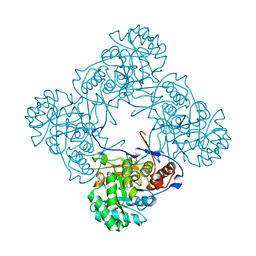 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5J4K
 
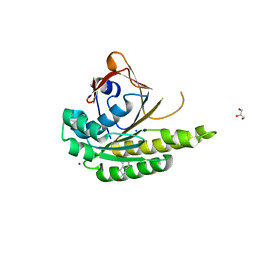 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4H
 
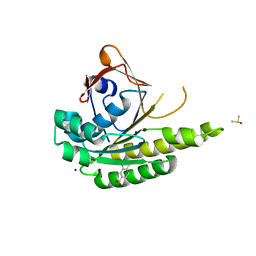 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4L
 
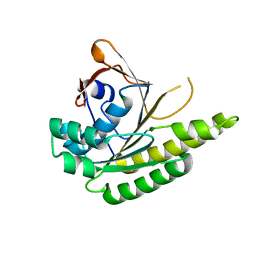 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
3KGV
 
 | |
5JED
 
 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
4B2L
 
 | | Humanised monomeric RadA in complex with L-methylester tryptophan | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION, methyl L-tryptophanate | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
1RQF
 
 | | Structure of CK2 beta subunit crystallized in the presence of a p21WAF1 peptide | | Descriptor: | Casein kinase II beta chain, Disordered segment of Cyclin-dependent kinase inhibitor 1, UNKNOWN, ... | | Authors: | Bertrand, L, Sayed, M.F, Pei, X.-Y, Parisini, E, Dhanaraj, V, Bolanos-Garcia, V.M, Allende, J.E, Blundell, T.L. | | Deposit date: | 2003-12-05 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the regulatory subunit of CK2 in the presence of a p21WAF1 peptide demonstrates flexibility of the acidic loop.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3W1G
 
 | | Crystal Structure of Human DNA ligase IV-Artemis Complex (Native) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Artemis-derived peptide, DNA ligase 4, ... | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
3W03
 
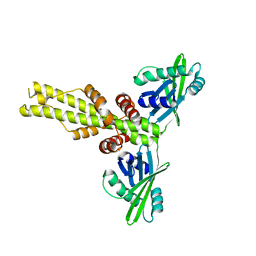 | | XLF-XRCC4 complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (8.492 Å) | | Cite: | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
3W1B
 
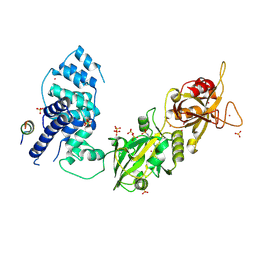 | |
5J5R
 
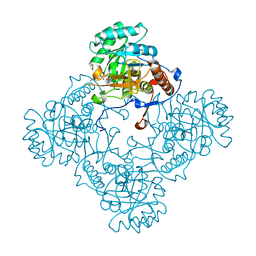 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor VCC234718 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, cyclohexyl{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}methanone | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-04-03 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Inosine Monophosphate Dehydrogenase, GuaB2, Is a Vulnerable New Bactericidal Drug Target for Tuberculosis.
ACS Infect Dis, 3, 2017
|
|
6YY6
 
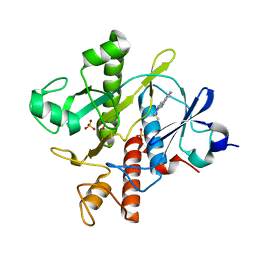 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-(1-ethylpyrazol-4-yl)pyrimidine-5-carbonitrile, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
