5F27
 
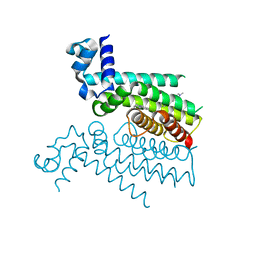 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 2 at 1.68A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{N}-methyl-1-(4-piperidin-1-ylphenyl)methanamine | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F04
 
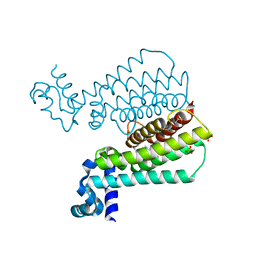 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 3 at 1.84A resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl-1-[3-[4-(methylaminomethyl)phenyl]-1,3-diazinan-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
4KL9
 
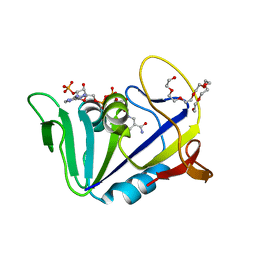 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in the space group C2 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KIJ
 
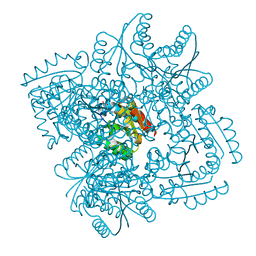 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinase dehydratase from Mycobacterium tuberculosis - compound 35c [3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid] | | Descriptor: | 3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KM2
 
 | |
5O08
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with Dephospho-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
4KM0
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with pyrimethamine | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
5O0D
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-Phenoxymandelic acid (Fragment 4) | | Descriptor: | (2~{S})-2-oxidanyl-2-(3-phenoxyphenyl)ethanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0H
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 2-(4-Chloro-3-nitrobenzoyl)benzoic acid (Fragment 6) | | Descriptor: | 2-(4-chloranyl-3-nitro-phenyl)carbonylbenzoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O06
 
 | | Crystal structure of APO form of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus | | Descriptor: | 1,2-ETHANEDIOL, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0B
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-Bromo-1H-indazole-5-carboxylic acid (Fragment 2) | | Descriptor: | 3-bromanyl-1~{H}-indazole-5-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
1EPO
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
4KLX
 
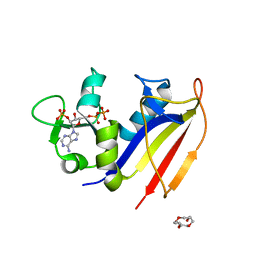 | |
4EFK
 
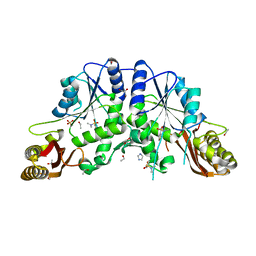 | | Pantothenate synthetase in complex with N,N-DIMETHYLTHIOPHENE-3-SULFONAMIDE | | Descriptor: | ETHANOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ciulli, A, Silvestre, H.L, Blundell, T.L, Abell, C. | | Deposit date: | 2012-03-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3H4T
 
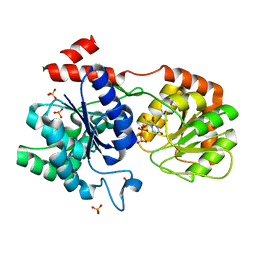 | | Chimeric Glycosyltransferase for the generation of novel natural products - GtfAH1 in complex with UDP-2F-Glc | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, PHOSPHATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
3H4I
 
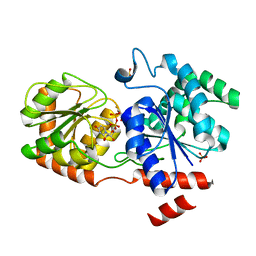 | | Chimeric Glycosyltransferase for the generation of novel natural products | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, SULFATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
4G5Y
 
 | | Crystal Structure of Mycobacterium tuberculosis Pantothenate synthetase in a ternary complex with ATP and N,N-DIMETHYLTHIOPHENE-3-SULFONAMIDE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Silvestre, H.L, Blundell, T.L, Abell, C. | | Deposit date: | 2012-07-18 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SAC
 
 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
3HWO
 
 | |
3ISJ
 
 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-methoxy-N-(methylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 5-methoxy-N-(methylsulfonyl)-1H-indole-2-carboxamide, ETHANOL, GLYCEROL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IMG
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.8 Ang resolution in a ternary complex with fragment compounds 5-methoxyindole and 1-benzofuran-2-carboxylic acid | | Descriptor: | 1-benzofuran-2-carboxylic acid, 5-methoxy-1H-indole, ETHANOL, ... | | Authors: | Ciulli, A, Hung, A.W, Silvestre, H.L, Wen, S, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4G5F
 
 | | Pantothenate synthetase in complex with racemate (2S)-2,3-DIHYDRO-1,4-BENZODIOXINE-2-CARBOXYLIC ACID and (2R)-2,3-DIHYDRO-1,4-BENZODIOXINE-2-CARBOXYLIC ACID | | Descriptor: | (2R)-2,3-dihydro-1,4-benzodioxine-2-carboxylic acid, (2S)-2,3-dihydro-1,4-benzodioxine-2-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SGF
 
 | | CRYSTAL STRUCTURE OF 7S NGF: A COMPLEX OF NERVE GROWTH FACTOR WITH FOUR BINDING PROTEINS (SERINE PROTEINASES) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NERVE GROWTH FACTOR, ... | | Authors: | Bax, B.D.V, Blundell, T.L, Murray-Rust, J, Mcdonald, N.Q. | | Deposit date: | 1997-08-08 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins.
Structure, 5, 1997
|
|
1IK9
 
 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
