6TUM
 
 | |
8BRQ
 
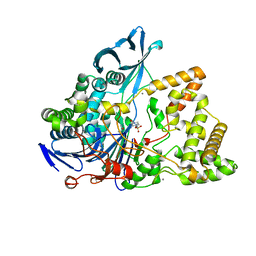 | | Crystal structure of a surface entropy reduction variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
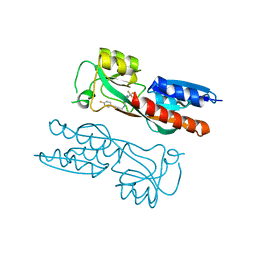
7NBW
 
 | |
6YHK
 
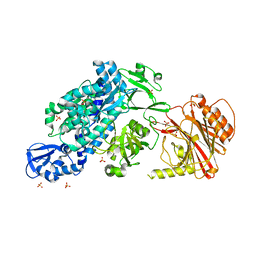 | | Crystal structure of full-length CNFy (C866S) from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, Cytotoxic necrotizing factor, SULFATE ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
8CIZ
 
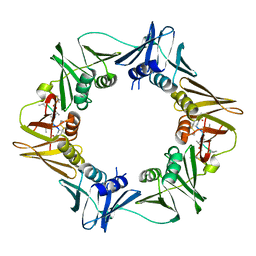 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Mycoplanecin A. | | Descriptor: | Beta sliding clamp, Mycoplanecin A | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
8CIY
 
 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Cyclohexyl-Griselimycin. | | Descriptor: | ACETATE ION, Beta sliding clamp, CALCIUM ION, ... | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
6YIZ
 
 | |
6YA1
 
 | | Zinc metalloprotease ProA | | Descriptor: | ACETATE ION, CALCIUM ION, ZINC ION, ... | | Authors: | Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2020-03-11 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Zinc metalloprotease ProA of Legionella pneumophila increases alveolar septal thickness in human lung tissue explants by collagen IV degradation.
Cell.Microbiol., 23, 2021
|
|
8BCI
 
 | |
8BCJ
 
 | |
8BRT
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
8BRS
 
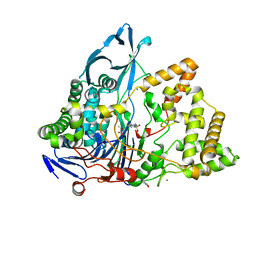 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
8BRR
 
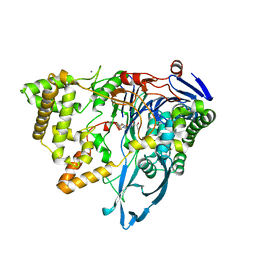 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
6RTE
 
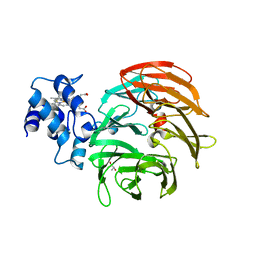 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
8BTJ
 
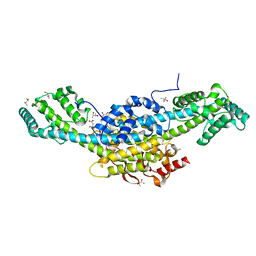 | | Murine cytomegalovirus protein M35 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MALONATE ION, Protein M35 | | Authors: | Schmelz, S, Van den Heuvel, J, Blankenfeldt, W. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Cytomegalovirus M35 Protein Directly Binds to the Interferon-beta Enhancer and Modulates Transcription of Ifnb1 and Other IRF3-Driven Genes.
J.Virol., 97, 2023
|
|
3JUM
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with 5-bromo-2-((1S,3R)-3-carboxycyclohexylamino)benzoic acid | | Descriptor: | 5-bromo-2-{[(1S,3R)-3-carboxycyclohexyl]amino}benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUQ
 
 | |
3JUN
 
 | |
3JUO
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3R)-piperidin-3-ylamino]benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUP
 
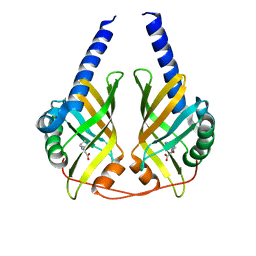 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (S)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3S)-piperidin-3-ylamino]benzoate, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1NXM
 
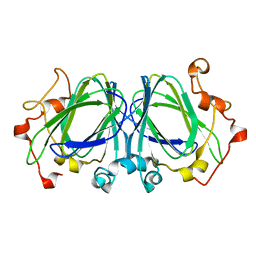 | | The high resolution structures of RmlC from Streptococcus suis | | Descriptor: | dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-11 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NYW
 
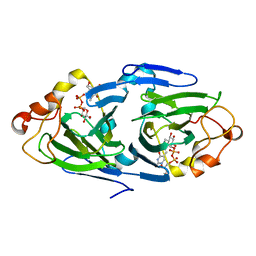 | | The high resolution structures of RmlC from Streptoccus suis in complex with dTDP-D-glucose | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NZC
 
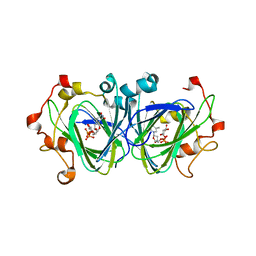 | | The high resolution structures of RmlC from Streptococcus suis in complex with dTDP-D-xylose | | Descriptor: | NICKEL (II) ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
8A26
 
 | | Lysophospholipase PlaA from Legionella pneumophila str. Corby - complex with palmitate | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Lysophospholipase A, MALONIC ACID, ... | | Authors: | Diwo, M.G, Blankenfeldt, W. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function relationships underpin disulfide loop cleavage-dependent activation of Legionella pneumophila lysophospholipase A PlaA.
Mol.Microbiol., 121, 2024
|
|
6RTD
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C, ... | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
