1DVH
 
 | | STRUCTURE AND DYNAMICS OF FERROCYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS STUDIED BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | 分子名称: | CYTOCHROME C553, HEME C | | 著者 | Blackledge, M.J, Medvedeva, S, Poncin, M, Guerlesquin, F, Bruschi, M, Marion, D. | | 登録日 | 1995-02-24 | | 公開日 | 1995-06-03 | | 最終更新日 | 2021-03-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
2NMQ
 
 | |
6OQQ
 
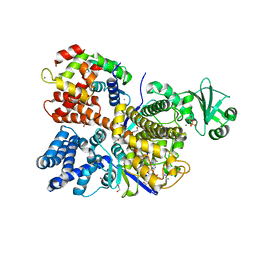 | | Legionella pneumophila SidJ/Saccharomyces cerevisiae calmodulin complex | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | 著者 | Tomchick, D.R, Tagliabracci, V.S, Black, M, Osinski, A. | | 登録日 | 2019-04-28 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases.
Science, 364, 2019
|
|
7MIS
 
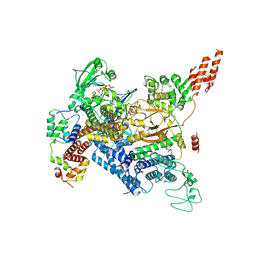 | | Cryo-EM structure of SidJ-SdeC-CaM reaction intermediate complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | 登録日 | 2021-04-17 | | 公開日 | 2021-08-18 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
7MIR
 
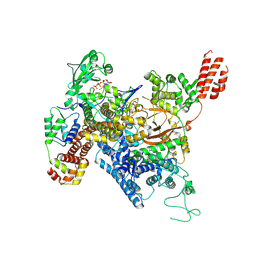 | | Cryo-EM structure of SidJ-SdeA-CaM reaction intermediate complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | 登録日 | 2021-04-17 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
1YSD
 
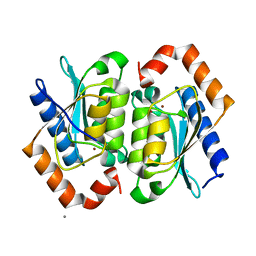 | | Yeast Cytosine Deaminase Double Mutant | | 分子名称: | CALCIUM ION, Cytosine deaminase, ZINC ION | | 著者 | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | 登録日 | 2005-02-08 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
1YSB
 
 | | Yeast Cytosine Deaminase Triple Mutant | | 分子名称: | CALCIUM ION, Cytosine deaminase, ZINC ION | | 著者 | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | 登録日 | 2005-02-08 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
1P6O
 
 | | The crystal structure of yeast cytosine deaminase bound to 4(R)-hydroxyl-3,4-dihydropyrimidine at 1.14 angstroms. | | 分子名称: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, ACETIC ACID, CALCIUM ION, ... | | 著者 | Ireton, G.C, Black, M.E, Stoddard, B.L. | | 登録日 | 2003-04-29 | | 公開日 | 2003-08-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | The 1.14 a crystal structure of yeast Cytosine deaminase. Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
Structure, 11, 2003
|
|
1OX7
 
 | |
1K70
 
 | | The Structure of Escherichia coli Cytosine Deaminase bound to 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-one | | 分子名称: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, Cytosine Deaminase, FE (III) ION | | 著者 | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | 登録日 | 2001-10-17 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K6W
 
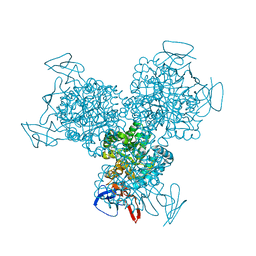 | | The Structure of Escherichia coli Cytosine Deaminase | | 分子名称: | Cytosine Deaminase, FE (III) ION | | 著者 | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | 登録日 | 2001-10-17 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1R9Y
 
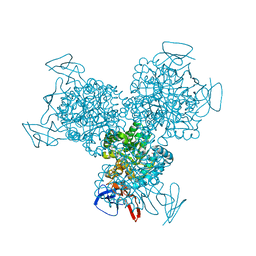 | | Bacterial cytosine deaminase D314A mutant. | | 分子名称: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1R9Z
 
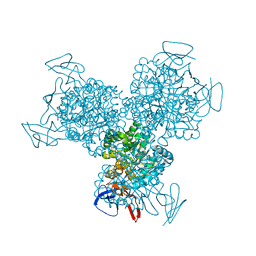 | | Bacterial cytosine deaminase D314S mutant. | | 分子名称: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RA5
 
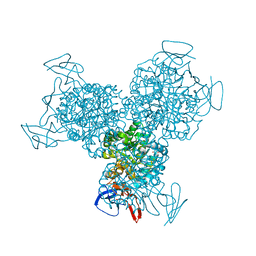 | | Bacterial cytosine deaminase D314A mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | 分子名称: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1R9X
 
 | | Bacterial cytosine deaminase D314G mutant. | | 分子名称: | Cytosine deaminase, FE (III) ION, GLYCEROL, ... | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RA0
 
 | | Bacterial cytosine deaminase D314G mutant bound to 5-fluoro-4-(S)-hydroxy-3,4-dihydropyrimidine. | | 分子名称: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RAK
 
 | | Bacterial cytosine deaminase D314S mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | 分子名称: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | 著者 | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
4HBU
 
 | | Crystal structure of CTX-M-15 extended-spectrum beta-lactamase in complex with avibactam (NXL104) | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Docquier, J.D, Benvenuti, M, Bruneau, J.M, Rossolini, G.M, Miossec, C, Black, M.T, Mangani, S. | | 登録日 | 2012-09-28 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-lactamases
Antimicrob.Agents Chemother., 57, 2013
|
|
4HBT
 
 | | Crystal structure of native CTX-M-15 extended-spectrum beta-lactamase | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Docquier, J.D, Benvenuti, M, Bruneau, J.M, Rossolini, G.M, Miossec, C, Black, M.T, Mangani, S. | | 登録日 | 2012-09-28 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-lactamases
Antimicrob.Agents Chemother., 57, 2013
|
|
4UX9
 
 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | 分子名称: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | 登録日 | 2014-08-20 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7PKU
 
 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | 分子名称: | 3C-like proteinase, Nucleoprotein | | 著者 | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | 登録日 | 2021-08-26 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
6TRI
 
 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | 分子名称: | CI, MOR, SULFATE ION | | 著者 | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | 登録日 | 2019-12-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.277 Å) | | 主引用文献 | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TO6
 
 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | 分子名称: | MOR | | 著者 | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | 登録日 | 2019-12-11 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4CO6
 
 | | Crystal structure of the Nipah virus RNA free nucleoprotein- phosphoprotein complex | | 分子名称: | BROMIDE ION, CHLORIDE ION, NUCLEOPROTEIN, ... | | 著者 | Yabukarksi, F, Lawrence, P, Tarbouriech, N, Bourhis, J.M, Jensen, M.R, Ruigrok, R.W.H, Blackledge, M, Volchkov, V, Jamin, M. | | 登録日 | 2014-01-27 | | 公開日 | 2014-08-13 | | 最終更新日 | 2014-09-17 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | Structure of Nipah Virus Unassembled Nucleoprotein in Complex with its Viral Chaperone.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6H5Q
 
 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | 分子名称: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | 著者 | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | 登録日 | 2018-07-25 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
