1HLA
 
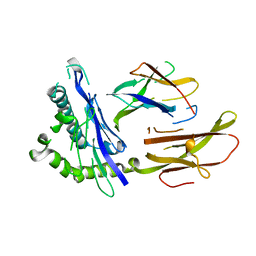 | | STRUCTURE OF THE HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN, HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2) (ALPHA CHAIN) | | Authors: | Bjorkman, P.J, Saper, M.A, Samraoui, B, Bennett, W.S, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1987-10-15 | | Release date: | 1988-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human class I histocompatibility antigen, HLA-A2.
Nature, 329, 1987
|
|
1CWV
 
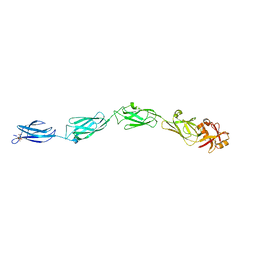 | |
2PZX
 
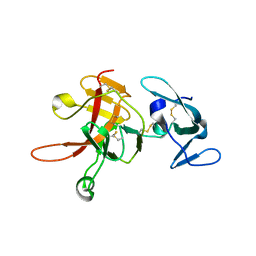 | | Structure of the methuselah ectodomain with peptide inhibitor | | Descriptor: | G-protein coupled receptor Mth | | Authors: | West Jr, A.P, Ja, W.W, Delker, S.L, Bjorkman, P.J, Benzer, S, Roberts, R.W. | | Deposit date: | 2007-05-18 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extension of Drosophila melanogaster life span with a GPCR peptide inhibitor.
Nat.Chem.Biol., 3, 2007
|
|
6MEG
 
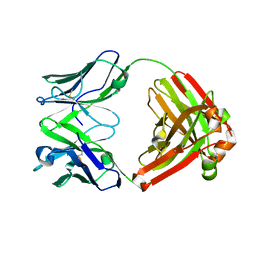 | | Crystal structure of human monoclonal antibody HEPC46 | | Descriptor: | antibody HEPC46 Heavy Chain, antibody HEPC46 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
6EDU
 
 | |
6XCM
 
 | |
6XCA
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C105 | | Descriptor: | C105 Heavy Chain, C105 Light Chain, SULFATE ION | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies.
Cell, 182, 2020
|
|
6XCN
 
 | |
8FYI
 
 | | Structure of HIV-1 BG505 SOSIP-HT1 in complex with one CD4 molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(6-8)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, C, Dam, K.A, Bjorkman, P.J. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Intermediate conformations of CD4-bound HIV-1 Env heterotrimers.
Nature, 623, 2023
|
|
8FYJ
 
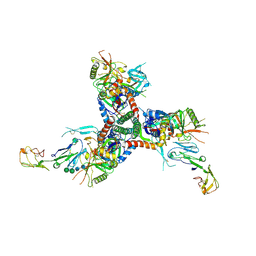 | | Structure of HIV-1 BG505 SOSIP-HT2 in complex with two CD4 molecules (class I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Fan, C, Dam, K.A, Bjorkman, P.J. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Intermediate conformations of CD4-bound HIV-1 Env heterotrimers.
Nature, 623, 2023
|
|
5FA2
 
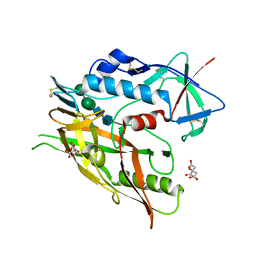 | | Crystal structure of 426c.TM4deltaV1-3 p120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, THIOCYANATE ION, ... | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
5FEC
 
 | |
5IGX
 
 | |
5I9Q
 
 | | Crystal structure of 3BNC55 Fab in complex with 426c.TM4deltaV1-3 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC55 Fab heavy chain, ... | | Authors: | Scharf, L, Chen, C, Bjorkman, P.J. | | Deposit date: | 2016-02-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
7M6F
 
 | |
7M6G
 
 | |
7M6I
 
 | |
7M6E
 
 | |
7M6H
 
 | |
7M6D
 
 | |
3HLA
 
 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN A2.1 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2.1) (ALPHA CHAIN) | | Authors: | Saper, M.A, Bjorkman, P.J, Wiley, D.C. | | Deposit date: | 1989-10-06 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution.
J.Mol.Biol., 219, 1991
|
|
7R8N
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C051 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8O
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C548 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8L
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody C099 and CR3022 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain, CR3022 Fab heavy chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8M
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C032 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
