3RPI
 
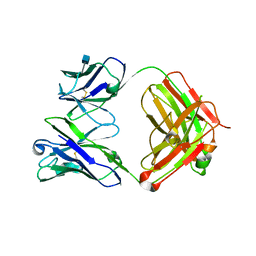 | | Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from highly potent anti-HIV neutralizing antibody, Light chain from highly potent anti-HIV neutralizing antibody | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Science, 333, 2011
|
|
1XED
 
 | |
7K90
 
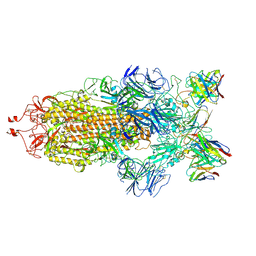 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C144 Fab Heavy Chain, C144 Fab Light Chain, ... | | Authors: | Barnes, C.O, Esswein, S.R, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
1OVZ
 
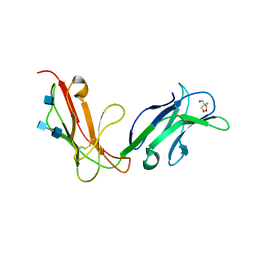 | | Crystal structure of human FcaRI | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Herr, A.B, Ballister, E.R, Bjorkman, P.J. | | Deposit date: | 2003-03-27 | | Release date: | 2003-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into IgA-mediated immune responses from the crystal structures of human Fc-alpha-RI and its complex with IgA1-Fc
Nature, 423, 2003
|
|
1P7Q
 
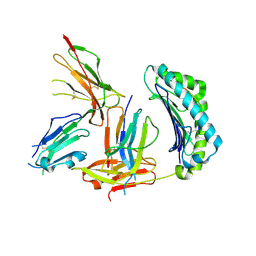 | | Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral MHC Receptor | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Willcox, B.E, Thomas, L.M, Bjorkman, P.J. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor.
Nat.Immunol., 4, 2003
|
|
5T3X
 
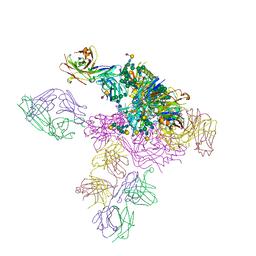 | |
1ZAG
 
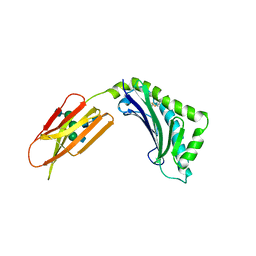 | | HUMAN ZINC-ALPHA-2-GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chirino, A.J, Sanchez, L.M, Bjorkman, P.J. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human ZAG, a fat-depleting factor related to MHC molecules.
Science, 283, 1999
|
|
5UEL
 
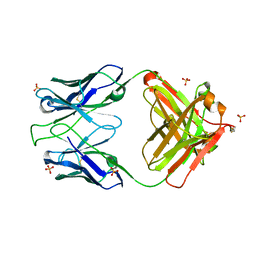 | | Crystal structure of 354NC102 Fab | | Descriptor: | 354NC102 Fab Heavy Chain, 354NC102 Fab Light Chain, SULFATE ION | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-02-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5D4K
 
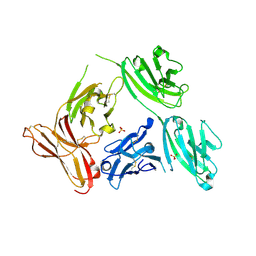 | |
1T7V
 
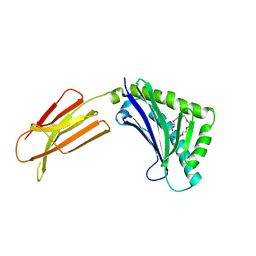 | | Zn-alpha-2-glycoprotein; baculo-ZAG PEG 200 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, ... | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
7K8N
 
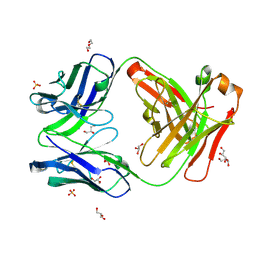 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C102 | | Descriptor: | C102 Fab Heavy Chain, C102 Fab Light Chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Q
 
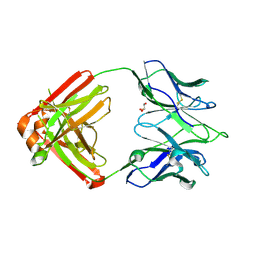 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Y
 
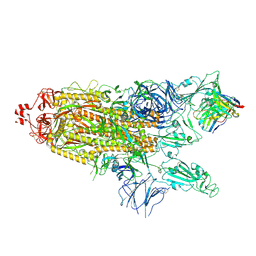 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8P
 
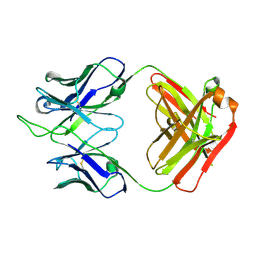 | |
7K8R
 
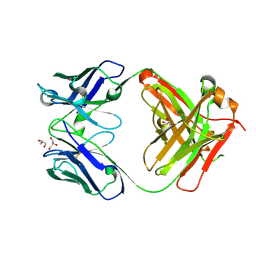 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C135 | | Descriptor: | C135 Fab Heavy Chain, C135 Fab Light Chain, GLYCEROL | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Z
 
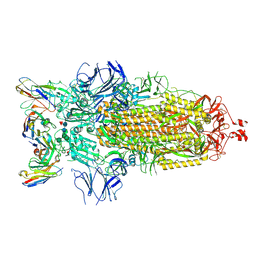 | |
7K8M
 
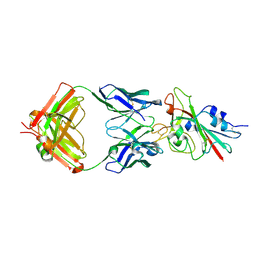 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8V
 
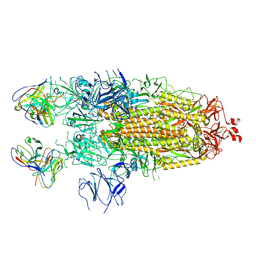 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | Authors: | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8O
 
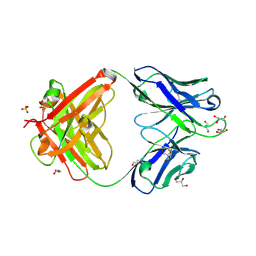 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8W
 
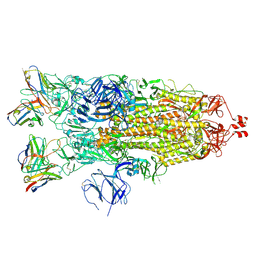 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C119 Fab Heavy Chain, ... | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7RYU
 
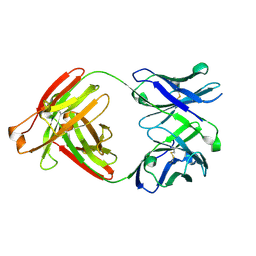 | |
5CJX
 
 | |
2GJ7
 
 | | Crystal Structure of a gE-gI/Fc complex | | Descriptor: | Glycoprotein E, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sprague, E.R, Wang, C, Baker, D, Bjorkman, P.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Crystal Structure of the HSV-1 Fc Receptor Bound to Fc Reveals a Mechanism for Antibody Bipolar Bridging.
Plos Biol., 4, 2006
|
|
2GIY
 
 | |
4Q6Y
 
 | | Crystal structure of a chemoenzymatic glycoengineered disialylated Fc (di-sFc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmed, A.A, Giddens, J, Pincetic, A, Lomino, J.V, Ravetch, J.V, Wang, L.X, Bjorkman, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of anti-inflammatory immunoglobulin g fc proteins.
J.Mol.Biol., 426, 2014
|
|
