3H76
 
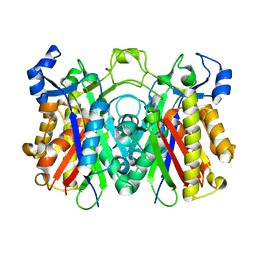 | | Crystal structure of PqsD, a key enzyme in Pseudomonas aeruginosa quinolone signal biosynthesis pathway | | 分子名称: | PQS biosynthetic enzyme | | 著者 | Bera, A.K, Atanasova, V, Parsons, J.F. | | 登録日 | 2009-04-24 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3HGV
 
 | |
6BP1
 
 | |
7MWR
 
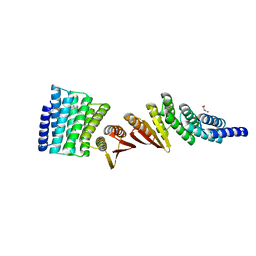 | | Structure of De Novo designed beta sheet heterodimer LHD101A53/B4 | | 分子名称: | LHD101A54, LHD101B4, MALONATE ION | | 著者 | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | 登録日 | 2021-05-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7MWQ
 
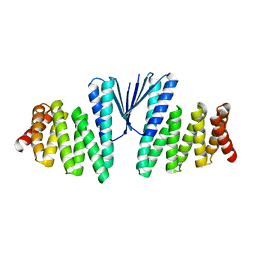 | | Structure of De Novo designed beta sheet heterodimer LHD29A53/B53 | | 分子名称: | LHD29A53, LHD29B53 | | 著者 | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | 登録日 | 2021-05-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
6BOF
 
 | |
6WHO
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHQ
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WI3
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | (SHA)W(DTH)DN(DSN)(DME)(DAS)K peptide macrocycle, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WMK
 
 | | Crystal structure of beta sheet heterodimer LHD29 | | 分子名称: | Beta sheet heterodimer LHD29 - Chain A, Beta sheet heterodimer LHD29 - Chain B | | 著者 | Bera, A.K, Sahtoe, D.D, Kang, A, Sankaran, B, Baker, D. | | 登録日 | 2020-04-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
6X9Z
 
 | |
6XI6
 
 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | 分子名称: | helical fusion design | | 著者 | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | 登録日 | 2020-06-19 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XH5
 
 | |
8E1E
 
 | |
8DT0
 
 | |
8EK4
 
 | |
8EOX
 
 | |
8EVM
 
 | |
8FIQ
 
 | |
8FVT
 
 | |
8FIN
 
 | |
8FIH
 
 | |
8FIT
 
 | |
