3FT8
 
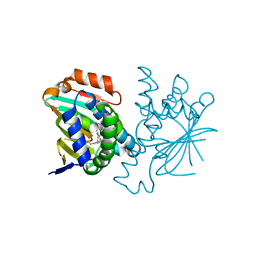 | | Structure of HSP90 bound with a noval fragment. | | Descriptor: | (5E,7S)-2-amino-7-(4-fluoro-2-pyridin-3-ylphenyl)-4-methyl-7,8-dihydroquinazolin-5(6H)-one oxime, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based identification of Hsp90 inhibitors.
Chemmedchem, 4, 2009
|
|
3FT5
 
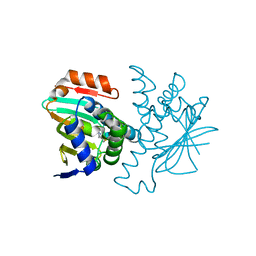 | | Structure of HSP90 bound with a novel fragment | | Descriptor: | 4-methyl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Identification of Hsp90 Inhibitors.
Chemmedchem, 4, 2009
|
|
3HZ1
 
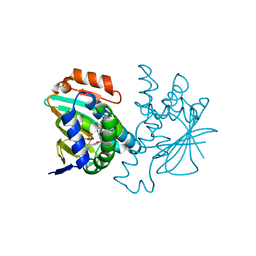 | | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03
TO BE PUBLISHED
|
|
3HYY
 
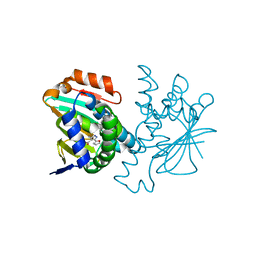 | | Crystal structure of Hsp90 with fragment 37-D04 | | Descriptor: | Heat shock protein HSP 90-alpha, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment 37-D04
TO BE PUBLISHED
|
|
3HYZ
 
 | | Crystal structure of Hsp90 with fragment 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragment 42-C03
TO BE PUBLISHED
|
|
3HZ5
 
 | | Crystal structure of Hsp90 with fragment Z064 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(5-furan-2-yl-3-methyl-1H-pyrazol-4-yl)butyl]-N-methyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment Z064
to be published
|
|
4N1B
 
 | | STRUCTURE OF KEAP1 KELCH DOMAIN WITH(1S,2R)-2-[(1S)-1-[(1-oxo-2,3-dihydro-1H-isoindol-2-Yl)methyl]-1,2,3,4-tetrahydroisoquinoline-2-Carbonyl]cyclohexane-1-carboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Smith, M.A, Duclos, S, Beaumont, E, Kwong, J, Brooks, M, Barker, J, Jnoff, E, Brookfield, F, Courade, J.P, Barker, O, Fryatt, T, Albrecht, C, Bromidge, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4L7D
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4L7C
 
 | | Structure of keap1 kelch domain with 2-{[(1S)-2-{[(1R,2S)-2-(1H-tetrazol-5-yl)cyclohexyl]carbonyl}-1,2,3,4-tetrahydroisoquinolin-1-yl]methyl}-1H-isoindole-1,3(2H)-dione | | Descriptor: | 2-{[(1S)-2-{[(1R,2S)-2-(1H-tetrazol-5-yl)cyclohexyl]carbonyl}-1,2,3,4-tetrahydroisoquinolin-1-yl]methyl}-1H-isoindole-1,3(2H)-dione, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4L7B
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
9CE4
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 indazole inhibitor compound 6 | | Descriptor: | (1S,4r)-4-[(1P)-5-chloro-1-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-6-yl]-1-(oxetan-3-yl)piperidin-1-ium, 1,2-ETHANEDIOL, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L, Zebisch, M, Henry, C, Barker, J.J. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery and Optimization of N-Heteroaryl Indazole LRRK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
7ZDD
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K10ac histone peptide. | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM33, Histone H3.X, ... | | Authors: | Caria, S, Duclos, S, Crespillo, S, Errey, J, Barker, J.J. | | Deposit date: | 2022-03-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Identification of Histone Peptide Binding Specificity and Small-Molecule Ligands for the TRIM33 alpha and TRIM33 beta Bromodomains.
Acs Chem.Biol., 17, 2022
|
|
6T8X
 
 | | Crystal structure of MAPKAPK2 (MK2) complexed with PF-3644022 and 5-(4-bromophenyl)-N-[4-(1-piperazinyl)phenyl]-N-(2-pyridinylmethyl)-2-furancarboxamide | | Descriptor: | (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one, 5-(4-bromophenyl)-~{N}-(4-piperazin-1-ylphenyl)-~{N}-(pyridin-2-ylmethyl)furan-2-carboxamide, CHLORIDE ION, ... | | Authors: | Beaumont, E.J, Barker, J. | | Deposit date: | 2019-10-25 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of MAPKAPK2 (MK2) complexed with PF-3644022 and 5-(4-bromophenyl)-N-[4-(1-piperazinyl)phenyl]-N-(2-pyridinylmethyl)-2-furancarboxamide
To Be Published
|
|
1SFI
 
 | | High resolution structure of a potent, cyclic protease inhibitor from sunflower seeds | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Luckett, S, Garcia, R.S, Barker, J.J, Konarev, A.V, Shewry, P, Clarke, A.R, Brady, R.L. | | Deposit date: | 1998-12-16 | | Release date: | 1999-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds.
J.Mol.Biol., 290, 1999
|
|
3L6C
 
 | | X-ray crystal structure of rat serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Wood, M, Schonfeld, D, Mather, O, Cesura, A, Barker, J. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
3L6R
 
 | | The structure of mammalian serine racemase: Evidence for conformational changes upon inhibitor binding | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Serine racemase | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Cesura, A, Felicetti, B, Barker, J. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-26 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
3L6B
 
 | | X-ray crystal structure of human serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Barker, J, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Cesura, A. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
8OUS
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 19 | | Descriptor: | (1S)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2-methyl-propan-1-ol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OUR
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 5-[1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)-2-propan-2-yl-imidazol-4-yl]-3-(trifluoromethyloxy)pyridin-2-amine, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OUT
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 22 | | Descriptor: | (1R)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-fluoranyl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2,2,2-tris(fluoranyl)ethanol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, Akkermans, O, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
7OJY
 
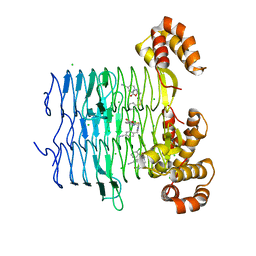 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 6 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-oxidanyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OKB
 
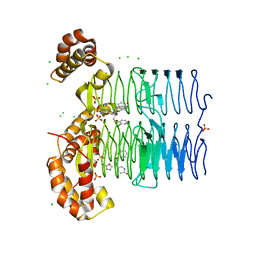 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 45 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OKC
 
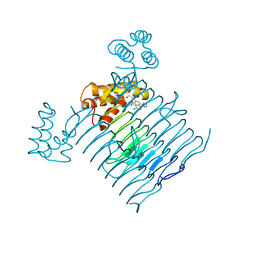 | | Crystal structure of Escherichia coli LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SODIUM ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OK1
 
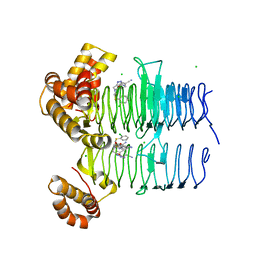 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJ6
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, ACETATE ION, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
