6HS6
 
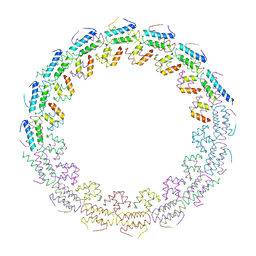 | | C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-09-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
7PQ0
 
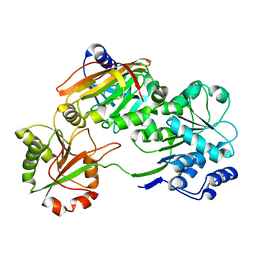 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PPZ
 
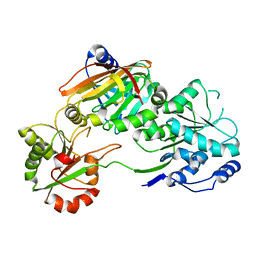 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
8Q1C
 
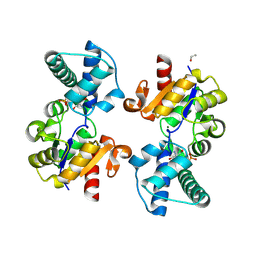 | | Substrate-free D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-phosphoglucomutase, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
8Q1F
 
 | | D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with native beta-glucose 1,6-bisphosphate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
8Q1D
 
 | | D10N variant of beta-phosphoglucomutase from Lactococcus lactis in complex with fructose 1,6-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, Beta-phosphoglucomutase, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
8Q1E
 
 | | D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with fructose 1,6-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, Beta-phosphoglucomutase, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
3TU8
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | BROMIDE ION, Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
3TUA
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) C94S mutant | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
7PZT
 
 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | Authors: | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
6RVU
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
6R1J
 
 | | Structure of the soluble AhlC triple head mutant of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2019-03-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
1ZUW
 
 | | Crystal structure of B.subtilis glutamate racemase (RacE) with D-Glu | | Descriptor: | D-GLUTAMIC ACID, glutamate racemase 1 | | Authors: | Ruzheinikov, S.N, Taal, M.A, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-Induced Conformational Changes in Bacillus subtilis Glutamate Racemase and Their Implications for Drug Discovery
Structure, 13, 2005
|
|
1BGV
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
5MHA
 
 | | D-2-hydroxyacid dehydrogenases (D2-HDH) from Haloferax mediterranei in complex with a mixture of 2-ketohexanoic acid and 2-hydroxyhexanoic acid, and NADPH (1.57 A resolution) | | Descriptor: | (2R)-2-hydroxyhexanoic acid, 1,2-ETHANEDIOL, 2-Ketohexanoic acid, ... | | Authors: | Bisson, C, Baker, P.J, Domenech Perez, J, Pramanpol, N, Harding, S.E, Rice, D.W, Ferrer, J. | | Deposit date: | 2016-11-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Productive ternary complexes of D-2-hydroxyacid dehydrogenase provide insights into the chiral specificity of its reaction mechanism
To Be Published
|
|
5MH5
 
 | | D-2-hydroxyacid dehydrogenases (D2-HDH) from Haloferax mediterranei in complex with 2-keto-hexanoic acid and NADP+ (1.4 A resolution) | | Descriptor: | 2-Ketohexanoic acid, D-2-hydroxyacid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Bisson, C, Baker, P.J, Domenech Perez, J, Pramanpol, N, Harding, S.E, Rice, D.W, Ferrer, J. | | Deposit date: | 2016-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Productive ternary complexes of D-2-hydroxyacid dehydrogenase provide insights into the chiral specificity of its reaction mechanism
To Be Published
|
|
5MH6
 
 | | D-2-hydroxyacid dehydrogenases (D2-HDH) from Haloferax mediterranei in complex with 2-ketohexanoic acid and NAD+ (1.35 A resolution) | | Descriptor: | 1,2-ETHANEDIOL, 2-Ketohexanoic acid, D-2-hydroxyacid dehydrogenase, ... | | Authors: | Bisson, C, Baker, P.J, Domenech Perez, J, Pramanpol, N, Harding, S.E, Rice, D.W, Ferrer, J. | | Deposit date: | 2016-11-23 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Productive ternary complexes of D-2-hydroxyacid dehydrogenase provide insights into the chiral specificity of its reaction mechanism
To Be Published
|
|
6G7C
 
 | | Nt2-CTD domains of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | Descriptor: | ImpA-related domain protein | | Authors: | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-04-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
6G7B
 
 | | Nt2 domain of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | Descriptor: | ImpA-related domain protein | | Authors: | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-04-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
6H2D
 
 | |
6GRJ
 
 | | Structure of the AhlB pore of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AhlB, CHLORIDE ION, ... | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2018-06-11 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
6H2E
 
 | |
6GRK
 
 | |
6H2F
 
 | | Structure of the pre-pore AhlB of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | AhlB, PHOSPHATE ION | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
6FWH
 
 | | Acanthamoeba IGPD in complex with R-C348 to 1.7A resolution | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Roberts, C.W, Bisson, C, Baker, P.J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional studies of histidine biosynthesis in Acanthamoeba spp. demonstrates a novel molecular arrangement and target for antimicrobials.
PLoS ONE, 13, 2018
|
|
