6Y1S
 
 | |
6Q8G
 
 | | Structure of Fucosylated D-antimicrobial peptide SB8 in complex with the Fucose-binding lectin PA-IIL at 1.190 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Q6X
 
 | | Structure of Fucosylated D-antimicrobial peptide SB6 in complex with the Fucose-binding lectin PA-IIL at 1.525 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Q79
 
 | | Structure of Fucosylated D-antimicrobial peptide SB4 in complex with the Fucose-binding lectin PA-IIL at 2.009 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6Y14
 
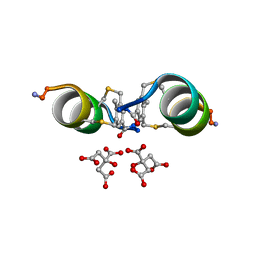 | |
6Y13
 
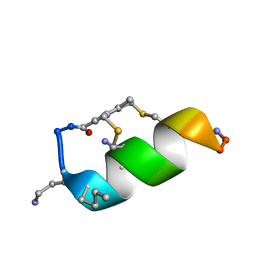 | |
3V3B
 
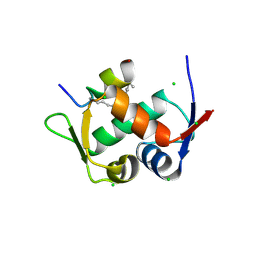 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
5XD0
 
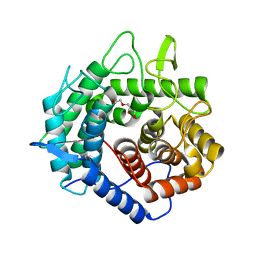 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
3V3V
 
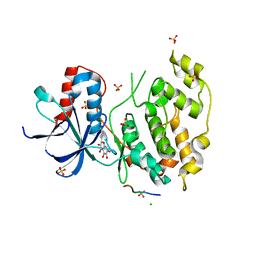 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
1QKG
 
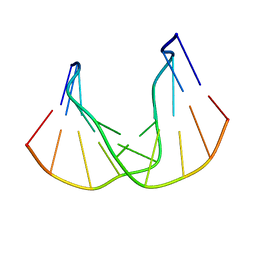 | | DNA DECAMER DUPLEX CONTAINING T-T DEWAR PHOTOPRODUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(HYD)TP*+TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Bae, S.-H, Choi, Y.-J, Choi, B.-S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-05-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Dewar Photoproduct of Thymidylyl(3'-->5')-Thymidine (Dewar Product) Exhibits Mutagenic Behavior in Accordance with the "A Rule".
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6TL8
 
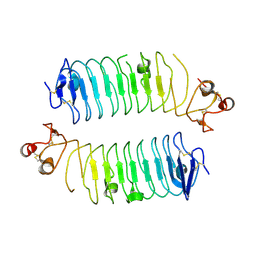 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | Authors: | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
2KAL
 
 | | NMR structure of fully methylated GATC site | | Descriptor: | 5'-D(*DCP*DGP*DCP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DCP*DGP*DC)-3', 5'-D(*DGP*DCP*DGP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DGP*DCP*DG)-3' | | Authors: | Bang, J, Bae, S, Park, C, Lee, J, Choi, B. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites.
J.Am.Chem.Soc., 130, 2008
|
|
1MFY
 
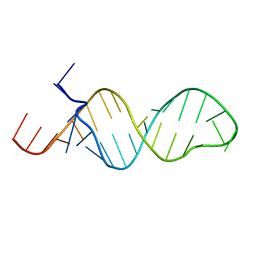 | | SOLUTION STRUCTURE OF INFLUENZA A VIRUS C4 PROMOTER | | Descriptor: | C4 promoter of influneza A virus | | Authors: | Lee, M.-K, Bae, S.-H, Park, C.-J, Cheong, H.-K, Cheong, C, Choi, B.-S. | | Deposit date: | 2002-08-14 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A single-nucleotide natural variation (U4 to C4) in an influenza A virus promoter exhibits a large structural change: implications for differential viral RNA synthesis by RNA-dependent RNA polymerase.
Nucleic Acids Res., 31, 2003
|
|
1M82
 
 | | SOLUTION STRUCTURE OF THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Descriptor: | RNA (25-MER): THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Authors: | Park, C.-J, Bae, S.-H, Lee, M.-K, Varani, G, Choi, B.-S. | | Deposit date: | 2002-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase
NUCLEIC ACIDS RES., 31, 2003
|
|
1P1A
 
 | | NMR structure of ubiquitin-like domain of hHR23B | | Descriptor: | UV excision repair protein RAD23 homolog B | | Authors: | Ryu, K.S, Lee, K.J, Bae, S.H, Kim, B.K, Kim, K.A, Choi, B.S. | | Deposit date: | 2003-04-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a
J.Biol.Chem., 278, 2003
|
|
5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7NEF
 
 | | Fucosylated linear peptide Fln65 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fln65, ... | | Authors: | Personne, H, Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
7NEW
 
 | | Fucosylated heterochiral linear peptide Fdln69 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 2.0 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Personne, H, Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
2JB5
 
 | | Fab fragment in complex with small molecule hapten, crystal form-1 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
2JB6
 
 | | Fab fragment in complex with small molecule hapten, crystal form-2 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
