4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1DAZ
 
 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE (RETROPEPSIN), N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1DG3
 
 | | STRUCTURE OF HUMAN GUANYLATE BINDING PROTEIN-1 IN NUCLEOTIDE FREE FORM | | Descriptor: | PROTEIN (INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1) | | Authors: | Prakash, B, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 1999-11-23 | | Release date: | 2000-10-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature, 403, 2000
|
|
8CB3
 
 | | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Papain-like protease nsp3 | | Authors: | Morin, B, Ferron, F, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid
To Be Published
|
|
1DUX
 
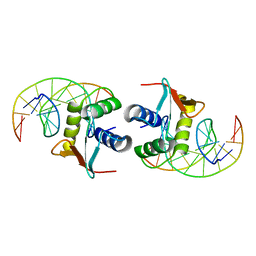 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
1DZL
 
 | |
1DW4
 
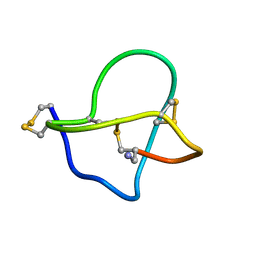 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1DXW
 
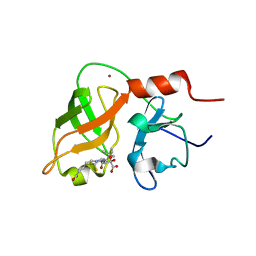 | | structure of hetero complex of non structural protein (NS) of hepatitis C virus (HCV) and synthetic peptidic compound | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Cordier, F, Narjes, F, Gerlach, B, Sambucini, S, Grzesiek, S, Matassa, V.G, Defrancesco, R, Bazzo, R. | | Deposit date: | 2000-01-17 | | Release date: | 2001-01-12 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Inhibitor Binding Induces Active Site Stabilisation of the Hcv Ns3 Protein Serine Protease Domain
Embo J., 19, 2000
|
|
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1DW6
 
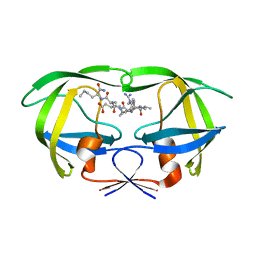 | | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease | | Descriptor: | HIV-1 PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Mahalingam, B, Louis, J.M, Reed, C.C, Adomat, J.M, Krouse, J, Wang, Y.F, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-01-24 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and kinetic analysis of drug resistant mutants of HIV-1 protease.
Eur.J.Biochem., 263, 1999
|
|
1DWO
 
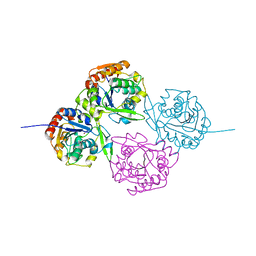 | | Crystal Structure of Hydroxynitrile Lyase from Manihot esculenta in Complex with Substrates Acetone and Chloroacetone:Implications for the Mechanism of Cyanogenesis | | Descriptor: | ACETONE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Forster, S, Miehlich, B, Wajant, H, Effenberger, F. | | Deposit date: | 1999-12-10 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Substrates Acetone and Chloroacetone: Implications for the Mechanism of Cyanogenesis
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1E1F
 
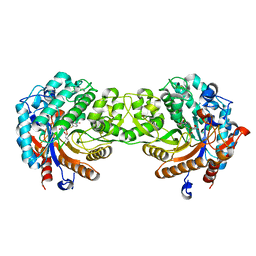 | | Crystal structure of a Monocot (Maize ZMGlu1) beta-glucosidase in complex with p-Nitrophenyl-beta-D-thioglucoside | | Descriptor: | 4-nitrophenyl 1-thio-beta-D-glucopyranoside, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-05-03 | | Release date: | 2001-02-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Monocotyledon (Maize Zmglu1) Beta-Glucosidase and a Model of its Complex with P-Nitrophenyl Beta-D-Thioglucoside
Biochem.J., 354, 2001
|
|
1DW5
 
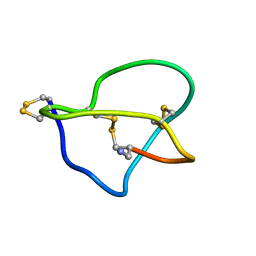 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: NO CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1E2S
 
 | | Crystal structure of an Arylsulfatase A mutant C69A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION, ... | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E3C
 
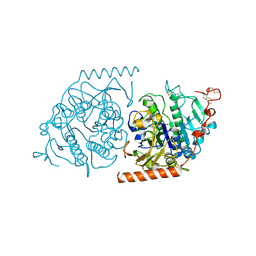 | | Crystal structure of an Arylsulfatase A mutant C69S soaked in synthetic substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-13 | | Release date: | 2001-03-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1DOB
 
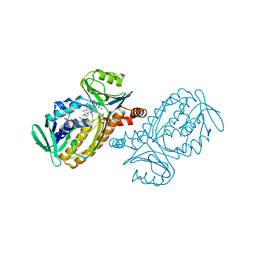 | | THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Palfey, B.A, Lah, M.S, Entsch, B, Massey, V, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mobile flavin of 4-OH benzoate hydroxylase.
Science, 266, 1994
|
|
1DZO
 
 | | Truncated PAK pilin from Pseudomonas aeruginosa | | Descriptor: | TYPE IV PILIN | | Authors: | Hazes, B, Read, R.J. | | Deposit date: | 2000-03-06 | | Release date: | 2000-06-11 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of Pseudomonas Aeruginosa Pak Pilin Suggests a Main-Chain-Dominated Mode of Receptor Binding
J.Mol.Biol., 299, 2000
|
|
1DUL
 
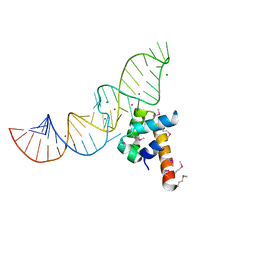 | | STRUCTURE OF THE RIBONUCLEOPROTEIN CORE OF THE E. COLI SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5 S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Rambo, R.P, Lucast, L, Rha, B, Doudna, J.A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-28 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonucleoprotein core of the signal recognition particle.
Science, 287, 2000
|
|
1E1Z
 
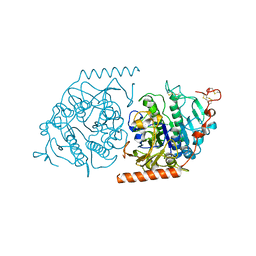 | | Crystal structure of an Arylsulfatase A mutant C69S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E15
 
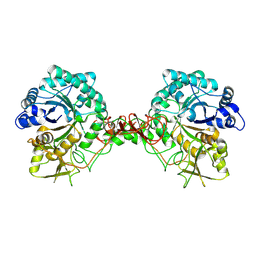 | | Chitinase B from Serratia Marcescens | | Descriptor: | CHITINASE B | | Authors: | Van Aalten, D.M.F, Synstad, B, Brurberg, M.B, Hough, E, Riise, B.W, Eijsink, V.G.H, Wierenga, R.K. | | Deposit date: | 2000-04-18 | | Release date: | 2000-08-18 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Two-Domain Chitotriosidase from Serratia Marcescens at 1.9 Angstrom Resoltuion
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E33
 
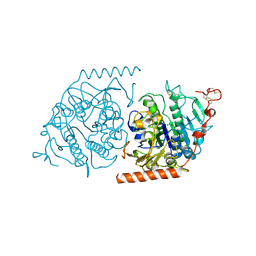 | | Crystal structure of an Arylsulfatase A mutant P426L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-06 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defective oligomerization of arylsulfatase a as a cause of its instability in lysosomes and metachromatic leukodystrophy.
J. Biol. Chem., 277, 2002
|
|
1DYQ
 
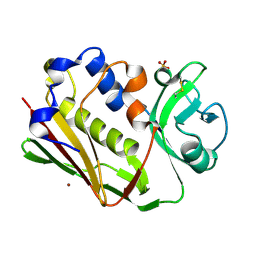 | | STAPHYLOCOCCAL ENTEROTOXIN A MUTANT VACCINE | | Descriptor: | Enterotoxin type A, SULFATE ION, ZINC ION | | Authors: | Krupka, H.I, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-02-05 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Abrogated Binding between Staphylococcal Enterotoxin a Superantigen Vaccine and Mhc-II?
Protein Sci., 11, 2002
|
|
1E0O
 
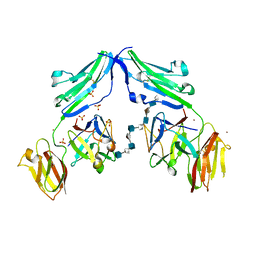 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
1DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
