5J2L
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | protein design 2L4HC2_11 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-29 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
8OW2
 
 | | Crystal structure of the p110alpha catalytic subunit from homo sapiens in complex with activator 1938 | | 分子名称: | 1-[7-[[2-[[4-(4-ethylpiperazin-1-yl)phenyl]amino]pyridin-4-yl]amino]-2,3-dihydroindol-1-yl]ethanone, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | 登録日 | 2023-04-26 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
1FMF
 
 | | REFINED SOLUTION STRUCTURE OF THE (13C,15N-LABELED) B12-BINDING SUBUNIT OF GLUTAMATE MUTASE FROM CLOSTRIDIUM TETANOMORPHUM | | 分子名称: | METHYLASPARTATE MUTASE S CHAIN | | 著者 | Hoffmann, B, Konrat, R, Tollinger, M, Huhta, M, Marsh, E.N.G, Kraeutler, B. | | 登録日 | 2000-08-17 | | 公開日 | 2002-02-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A protein pre-organized to trap the nucleotide moiety of coenzyme B(12): refined solution structure of the B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum.
Chembiochem, 2, 2001
|
|
1FD9
 
 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | 分子名称: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | 著者 | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | 登録日 | 2000-07-20 | | 公開日 | 2001-07-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
5L33
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | denovo NTF2 | | 著者 | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-08-03 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5KKM
 
 | | Con-Vc11-22 | | 分子名称: | O2_contryphan_Vc1 prepropeptide | | 著者 | Chittoor, B, Krishnarjuna, B, MacRaild, C.A, Robinson, S.D. | | 登録日 | 2016-06-22 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Single Disulfide-Directed beta-Hairpin Fold. Dynamics, Stability, and Engineering.
Biochemistry, 56, 2017
|
|
1GAI
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE | | 分子名称: | 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | 著者 | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | 登録日 | 1996-03-06 | | 公開日 | 1996-08-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
7TD6
 
 | | aRML prion fibril | | 分子名称: | Major prion protein | | 著者 | Hoyt, F, Standke, H.G, Artikis, E, Caughey, B, Kraus, A. | | 登録日 | 2021-12-30 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure of anchorless RML prion reveals variations in shared motifs between distinct strains.
Nat Commun, 13, 2022
|
|
3A3T
 
 | | The oxidoreductase NmDsbA1 from N. meningitidis | | 分子名称: | Thiol:disulfide interchange protein DsbA | | 著者 | Vivian, J.P, Scoullar, J, Bushell, S, Beddoe, T, Wilce, M.C.J, Byres, E, Boyle, T.P, Rimmer, K, Doak, B, Simpson, J.S, Graham, B, Heras, B, Kahler, C.M, Rossjohn, J, Scanlon, M.J. | | 登録日 | 2009-06-19 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis
J.Mol.Biol., 394, 2009
|
|
1IBA
 
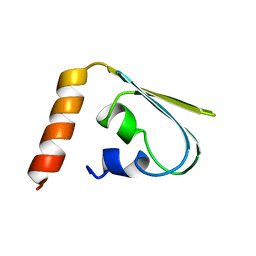 | | GLUCOSE PERMEASE (DOMAIN IIB), NMR, 11 STRUCTURES | | 分子名称: | GLUCOSE PERMEASE | | 著者 | Eberstadt, M, Grdadolnik, S.G, Gemmecker, G, Kessler, H, Buhr, A, Erni, B. | | 登録日 | 1996-03-23 | | 公開日 | 1996-10-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the IIB domain of the glucose transporter of Escherichia coli.
Biochemistry, 35, 1996
|
|
6FER
 
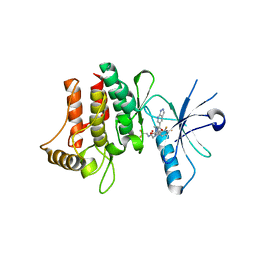 | | Crystal Structure of human DDR2 kinase in complex with 2-[4,5-difluoro-2-oxo-1'-(1H-pyrazolo[3,4-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide | | 分子名称: | 2-[4,5-bis(fluoranyl)-2-oxidanylidene-1'-(1~{H}-pyrazolo[3,4-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Discoidin domain-containing receptor 2 | | 著者 | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | 登録日 | 2018-01-03 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6NZ2
 
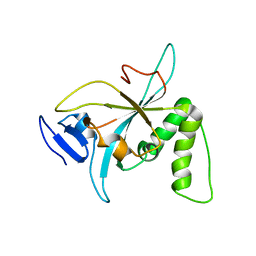 | |
5D7C
 
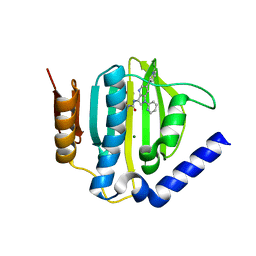 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D7R
 
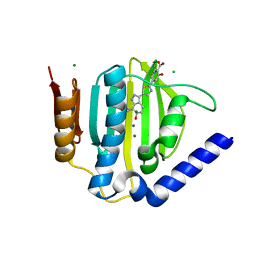 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-14 | | 公開日 | 2015-11-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
8CQY
 
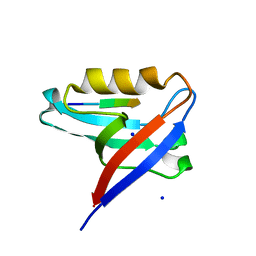 | | Crystal structure of the PTPN3 PDZ domain bound to the PBM TACE C-terminal peptide | | 分子名称: | Disintegrin and metalloproteinase domain-containing protein 17, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | 著者 | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | 登録日 | 2023-03-07 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-05-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
5DK6
 
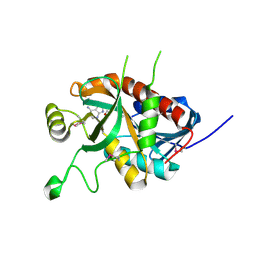 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | 分子名称: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | 著者 | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2015-09-03 | | 公開日 | 2015-11-04 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
6FY0
 
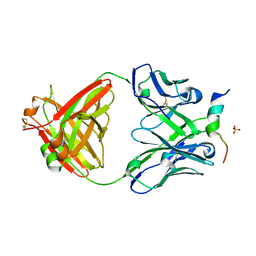 | | Crystal structure of a V2-directed, RV144 vaccine-like antibody from HIV-1 infection, CAP228-16H, bound to a heterologous V2 peptide | | 分子名称: | CAP228-16H Heavy Chain, CAP228-16H Light Chain, CAP45 V2 peptide, ... | | 著者 | Wibmer, C.K, Fernandes, M, Vijayakumar, B, Dirr, H.W, Sayed, Y, Moore, P.L, Morris, L. | | 登録日 | 2018-03-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | V2-Directed Vaccine-like Antibodies from HIV-1 Infection Identify an Additional K169-Binding Light Chain Motif with Broad ADCC Activity.
Cell Rep, 25, 2018
|
|
8BN8
 
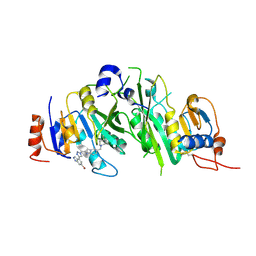 | | METTL3-METTL14 heterodimer bound to the SAM competitive small molecule inhibitor STM3006 | | 分子名称: | 2-[[4-(6-bromanyl-2~{H}-indazol-4-yl)-1,2,3-triazol-1-yl]methyl]-6-[(4,4-dimethylpiperidin-1-yl)methyl]imidazo[1,2-a]pyridine, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit | | 著者 | Pilka, E.S, Thomas, B, Blackaby, W, Hardick, D, Feeney, K, Ridgill, M, Rotty, B, Rausch, O. | | 登録日 | 2022-11-13 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.213 Å) | | 主引用文献 | Inhibition of METTL3 Results in a Cell-Intrinsic Interferon Response That Enhances Antitumor Immunity.
Cancer Discov, 13, 2023
|
|
5HIX
 
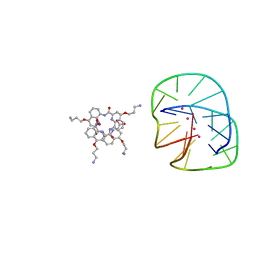 | | Cocrystal structure of an anti-parallel DNA G-quadruplex and a tetra-Quinoline Foldamer | | 分子名称: | 4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-nitroquinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinoline-2-carboxylic acid, Dimeric G-quadruplex, POTASSIUM ION | | 著者 | Mandal, P.K, Baptiste, B, Langlois d'Estaintot, B, Kauffmann, B, Huc, I. | | 登録日 | 2016-01-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State.
Chembiochem, 17, 2016
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D7D
 
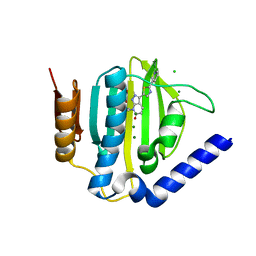 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
6WI6
 
 | | Crystal structure of plantacyclin B21AG | | 分子名称: | MALONATE ION, Plantacyclin B21AG | | 著者 | Smith, A.T, Gor, M.C, Vezina, B, McMahon, R, King, G, Panjikar, S, Rehm, B, Martin, J. | | 登録日 | 2020-04-08 | | 公開日 | 2021-01-06 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and site-directed mutagenesis of circular bacteriocin plantacyclin B21AG reveals cationic and aromatic residues important for antimicrobial activity.
Sci Rep, 10, 2020
|
|
