6CN5
 
 | |
6FY0
 
 | | Crystal structure of a V2-directed, RV144 vaccine-like antibody from HIV-1 infection, CAP228-16H, bound to a heterologous V2 peptide | | Descriptor: | CAP228-16H Heavy Chain, CAP228-16H Light Chain, CAP45 V2 peptide, ... | | Authors: | Wibmer, C.K, Fernandes, M, Vijayakumar, B, Dirr, H.W, Sayed, Y, Moore, P.L, Morris, L. | | Deposit date: | 2018-03-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | V2-Directed Vaccine-like Antibodies from HIV-1 Infection Identify an Additional K169-Binding Light Chain Motif with Broad ADCC Activity.
Cell Rep, 25, 2018
|
|
6N48
 
 | | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Liu, X, Masoudi, A, Kahsai, A.W, Huang, L.Y, Pani, B, Hirata, K, Ahn, S, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of beta2AR regulation by an intracellular positive allosteric modulator.
Science, 364, 2019
|
|
6NE5
 
 | |
6CN6
 
 | | RORC2 LBD complexed with compound 34 | | Descriptor: | 3-cyano-N-{3-[1-(cyclopentanecarbonyl)piperidin-4-yl]-1,4-dimethyl-1H-indol-5-yl}benzamide, Nuclear receptor ROR-gamma | | Authors: | Kauppi, B, Vajdos, F. | | Deposit date: | 2018-03-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 3-Cyano- N-(3-(1-isobutyrylpiperidin-4-yl)-1-methyl-4-(trifluoromethyl)-1 H-pyrrolo[2,3- b]pyridin-5-yl)benzamide: A Potent, Selective, and Orally Bioavailable Retinoic Acid Receptor-Related Orphan Receptor C2 Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
6CT2
 
 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZUX
 
 | |
4UB7
 
 | | High-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) showing an extreme distortion of the ATP-binding loop combined with a pi-halogen bond | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
4UAO
 
 | |
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
8BN8
 
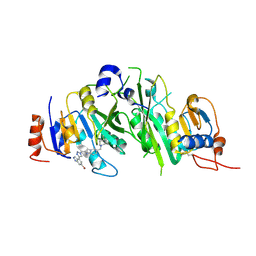 | | METTL3-METTL14 heterodimer bound to the SAM competitive small molecule inhibitor STM3006 | | Descriptor: | 2-[[4-(6-bromanyl-2~{H}-indazol-4-yl)-1,2,3-triazol-1-yl]methyl]-6-[(4,4-dimethylpiperidin-1-yl)methyl]imidazo[1,2-a]pyridine, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit | | Authors: | Pilka, E.S, Thomas, B, Blackaby, W, Hardick, D, Feeney, K, Ridgill, M, Rotty, B, Rausch, O. | | Deposit date: | 2022-11-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Inhibition of METTL3 Results in a Cell-Intrinsic Interferon Response That Enhances Antitumor Immunity.
Cancer Discov, 13, 2023
|
|
8FU3
 
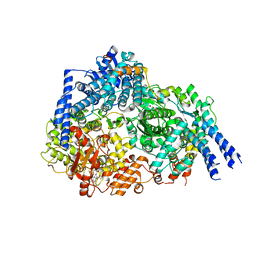 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
4UBA
 
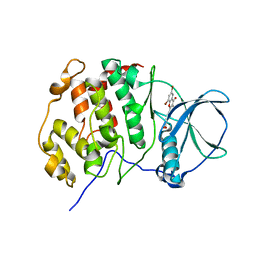 | | Low-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
6O8B
 
 | | Crystal structure of STING CTD in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6Z9M
 
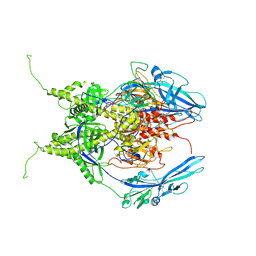 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | Descriptor: | Envelope glycoprotein B | | Authors: | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
8G7B
 
 | |
8G70
 
 | | SARS-CoV-2 spike/nanobody mixture complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 spike/nanobody mixture complex
To Be Published
|
|
5M5B
 
 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
8G7A
 
 | |
8G78
 
 | | Local refinement of SARS-CoV-2 spike/nanobody mixture complex around NTD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, Nanosota-6, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Local refinement of SARS-CoV-2 spike/nanobody mixture complex around NTD
To Be Published
|
|
8G7C
 
 | |
6O8C
 
 | | Crystal structure of STING CTT in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6XEZ
 
 | | Structure of SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Llewellyn, E.C, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex.
Cell, 182, 2020
|
|
2V4L
 
 | | complex of human phosphoinositide 3-kinase catalytic subunit gamma (p110 gamma) with PIK-284 | | Descriptor: | 3-[4-AMINO-1-(1-METHYLETHYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL]PHENOL, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Apsel, B, Gonzalez, B, Blair, J.A, Nazif, T.M, Feldman, M.E, Williams, R.L, Shokat, K.M, Knight, Z.A. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeted Polypharmacology: Discovery of Dual Inhibitors of Tyrosine and Phosphoinositide Kinases.
Nat.Chem.Biol., 4, 2008
|
|
