6E58
 
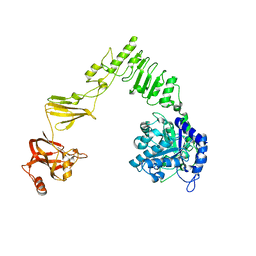 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) | | Descriptor: | CALCIUM ION, Secreted Endo-beta-N-acetylglucosaminidase (EndoS) | | Authors: | Klontz, E.H, Trastoy, B, Gunther, S, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6LIH
 
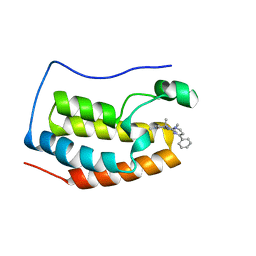 | | BRD4 BD1 bound with compound 10 | | Descriptor: | (3~{R})-1,3-dimethyl-6-[(4-phenylpyrimidin-2-yl)amino]-4-propan-2-yl-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62030542 Å) | | Cite: | BRD4 BD1 bound with compound 10
To Be Published
|
|
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MO1
 
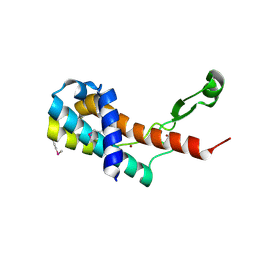 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | Descriptor: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
6LJI
 
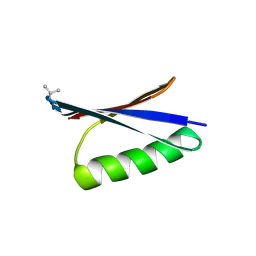 | | X-ray structure of synthetic GB1 domain with mutations K10(DVA), T11V | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Penmatsa, A, Chatterjee, J, Majumder, P, Khatri, B. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Increasing protein stability by engineering the n -> pi * interaction at the beta-turn.
Chem Sci, 11, 2020
|
|
4MRW
 
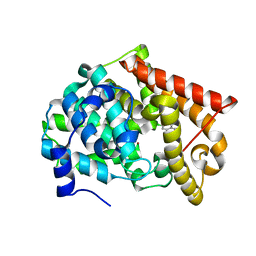 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | Descriptor: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
6ELG
 
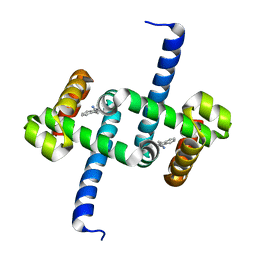 | | Tryptophan Repressor TrpR from E.coli variant M42F T44L T81I S88Y with Indole-3-acetonitrile | | Descriptor: | 1H-indol-3-ylacetonitrile, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
6EM9
 
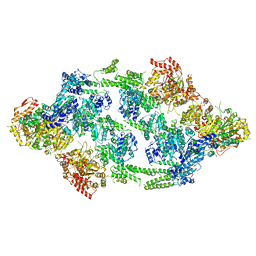 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6LIM
 
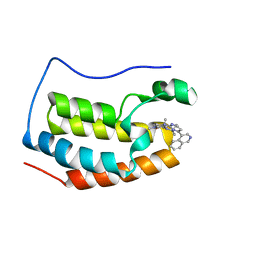 | | BRD4-BD1 bound with compound 40 | | Descriptor: | (3~{R})-6-[(4-isoquinolin-4-ylpyrimidin-2-yl)amino]-1,3-dimethyl-4-propan-2-yl-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75909507 Å) | | Cite: | BRD4-BD1 bound with compound 40
To Be Published
|
|
4MMH
 
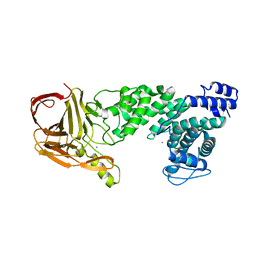 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
6E7I
 
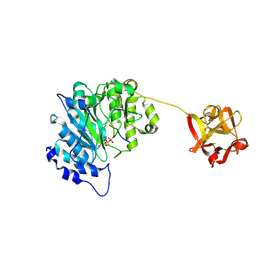 | | Human ppGalNAcT2 I253A/L310A Mutant with EA2 and UDP | | Descriptor: | EA2, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, ... | | Authors: | Bertozzi, C.R, Schumann, B, Agbay, A.J. | | Deposit date: | 2018-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
6EHF
 
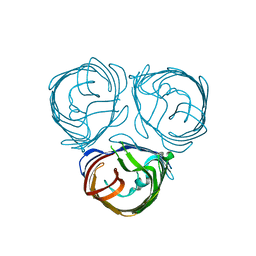 | |
6E9C
 
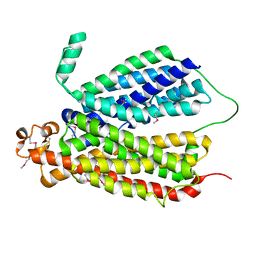 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
4MMI
 
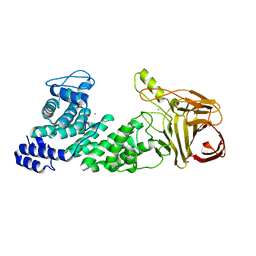 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
6EK5
 
 | |
6EEG
 
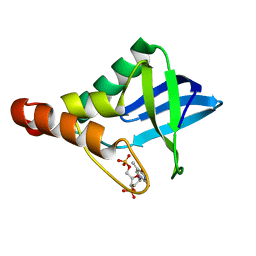 | |
6EN3
 
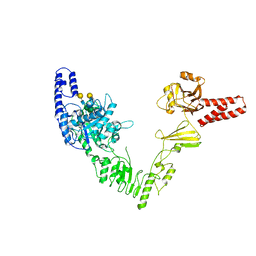 | | Crystal structure of full length EndoS from Streptococcus pyogenes in complex with G2 oligosaccharide. | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2,Multifunctional-autoprocessing repeats-in-toxin, NICKEL (II) ION, ... | | Authors: | Trastoy, B, Klontz, E.H, Orwenyo, J, Marina, A, Wang, L.X, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S.
Nat Commun, 9, 2018
|
|
4MOU
 
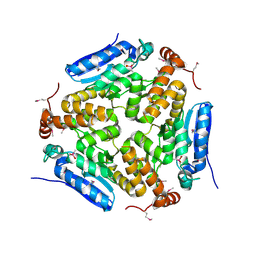 | | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282 | | Descriptor: | CACODYLATE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Chapman, H.C, Cooper, D.R, Geffken, K.T, Cymborowski, M.T, Osinski, T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282
To be Published
|
|
6ENI
 
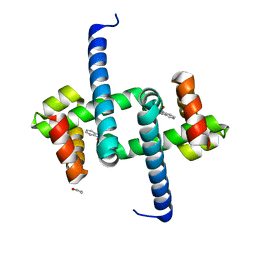 | | Tryptophan Repressor TrpR from E.coli variant T44L S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-10-04 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
6L6V
 
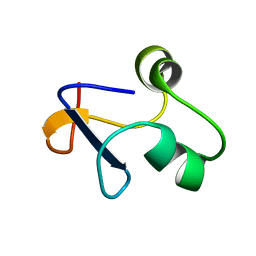 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
6ENQ
 
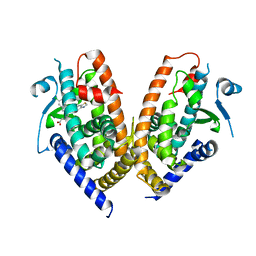 | | Structure of human PPAR gamma LBD in complex with Lanifibranor (IVA337) | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Boubia, B, Poupardin, O, Barth, M, Amaudrut, J, Broqua, P, Tallandier, M, Zeyer, D. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, Synthesis, and Evaluation of a Novel Series of Indole Sulfonamide Peroxisome Proliferator Activated Receptor (PPAR) alpha / gamma / delta Triple Activators: Discovery of Lanifibranor, a New Antifibrotic Clinical Candidate.
J. Med. Chem., 61, 2018
|
|
4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | Descriptor: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
6EHE
 
 | |
6EO7
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
4MVN
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
