5WKQ
 
 | | 2.10 A resolution structure of IpaB (residues 74-242) from Shigella flexneri | | Descriptor: | Invasin IpaB | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.L, Picking, W.D. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB.
Protein Sci., 27, 2018
|
|
3L5A
 
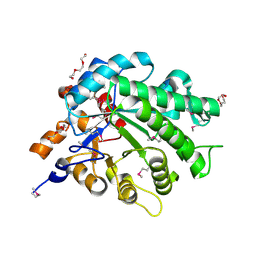 | | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus | | Descriptor: | NADH/flavin oxidoreductase/NADH oxidase, TRIETHYLENE GLYCOL | | Authors: | Lam, R, Gordon, R.D, Vodsedalek, J, Battaile, K.P, Grebemeskel, S, Lam, K, Romanov, V, Chan, T, Mihajlovic, V, Thompson, C.M, Guthrie, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus
To be Published
|
|
3KWL
 
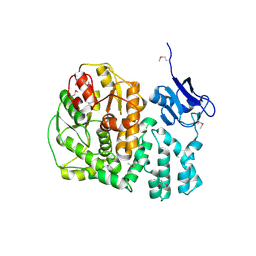 | | Crystal structure of a hypothetical protein from Helicobacter pylori | | Descriptor: | uncharacterized protein | | Authors: | Lam, R, Thompson, C.M, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a hypothetical protein from Helicobacter pylori
To be Published
|
|
5C98
 
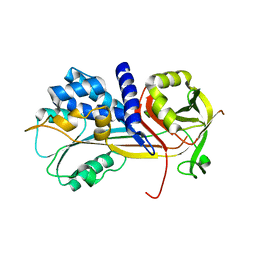 | | 1.45A resolution structure of SRPN18 from Anopheles gambiae | | Descriptor: | AGAP007691-PB | | Authors: | Lovell, S, Battaile, K.P, Gulley, M, Zhang, X, Meekins, D.A, Gao, F.P, Michel, K. | | Deposit date: | 2015-06-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 angstrom resolution structure of SRPN18 from the malaria vector Anopheles gambiae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3Q8C
 
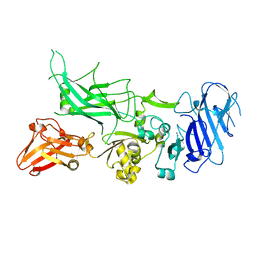 | | Crystal structure of Protective Antigen W346F (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8A
 
 | | Crystal structure of WT Protective Antigen (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Rajapaksha, M, Lovell, S, Janowiak, B.E, Andra, K.K, Battaile, K.P, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3MHZ
 
 | | 1.7A structure of 2-fluorohistidine labeled Protective Antigen | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Wimalasena, D.S, Janowiak, B.E, Miyagi, M, Sun, J, Hajduch, J, Pooput, C, Kirk, K.L, Bann, J.G. | | Deposit date: | 2010-04-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation.
Biochemistry, 49, 2010
|
|
3K2A
 
 | | Crystal structure of the homeobox domain of human homeobox protein Meis2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Homeobox protein Meis2 | | Authors: | Lam, R, Soloveychik, M, Battaile, K.P, Romanov, V, Lam, K, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the homeobox domain of human homeobox protein Meis2
To be Published
|
|
3Q8F
 
 | | Crystal structure of 2-Fluorohistine labeled Protective Antigen (pH 5.8) | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
5FBM
 
 | | Crystal Structure of Histone Like Protein (HLP) from Streptococcus mutans Refined to 1.9 A Resolution | | Descriptor: | DNA-binding protein HU | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, O'Neil, P, Biswas, I. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of histone-like protein from Streptococcus mutans refined to 1.9 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3Q8B
 
 | | Crystal structure of WT Protective Antigen (pH 9.0) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3R9I
 
 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3Q8E
 
 | | Crystal structure of Protective Antigen W346F (pH 8.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
4HIN
 
 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | Descriptor: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
4MLK
 
 | | 3.05A resolution structure of CT584 from Chlamydia trachomatis | | Descriptor: | CT584 protein | | Authors: | Hickey, J, Lovell, S, Kemege, K, Barta, M.L, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2013-09-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Structure of CT584 from Chlamydia trachomatis refined to 3.05 angstrom resolution.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5IXE
 
 | | 1.75A RESOLUTION STRUCTURE OF 5-Fluoroindole BOUND BETA-GLYCOSIDASE (W33G) FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-fluoro-1H-indole, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Full and Partial Agonism of a Designed Enzyme Switch.
ACS Synth Biol, 5, 2016
|
|
8G0G
 
 | | Crystal structure of diphtheria toxin H223Q/H257Q double mutant (pH 4.5) | | Descriptor: | ADENYLYL-3'-5'-PHOSPHO-URIDINE-3'-MONOPHOSPHATE, Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Ladokhin, A.S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine Protonation and Conformational Switching in Diphtheria Toxin Translocation Domain.
Toxins, 15, 2023
|
|
4NWG
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation
To be Published
|
|
3P8K
 
 | | Crystal Structure of a putative carbon-nitrogen family hydrolase from Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gordon, R.D, Qiu, W, Battaile, K, Lam, K, Soloveychik, M, Benetteraj, D, Romanov, V, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-10-14 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of carbon-nitrogen family hydrolase from Staphylococcus aureus
To be Published
|
|
6AW8
 
 | | 2.25A resolution domain swapped dimer structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
4NWF
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation
To be Published
|
|
6AW9
 
 | | 2.55A resolution structure of SAH bound catechol O-methyltransferase (COMT) L136M from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
4TJW
 
 | | Crystal Structure of human Tankyrase 2 in complex with PJ-34. | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKG
 
 | | Crystal Structure of human Tankyrase 2 in complex with AZD2281. | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TK5
 
 | | Crystal Structure of human Tankyrase 2 in complex with EB47. | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
