5NMU
 
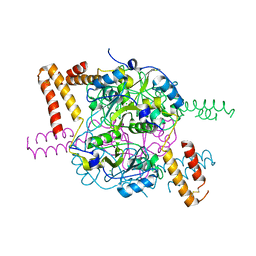 | | Structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria | | Descriptor: | CBS-CP12, CHLORIDE ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4A7T
 
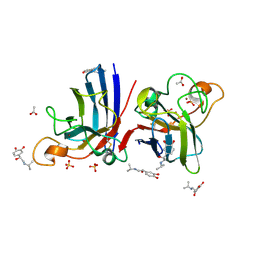 | | Structure of human I113T SOD1 mutant complexed with isoproteranol in the p21 space group | | Descriptor: | ACETATE ION, COPPER (II) ION, ISOPRENALINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A7Q
 
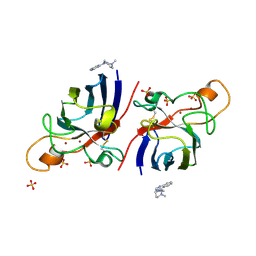 | | Structure of human I113T SOD1 mutant complexed with 4-(4-methyl-1,4- diazepan-1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-Ray Crystallography and Computational Docking for the Detection and Development of Protein-Ligand Interactions.
Curr.Med.Chem., 20, 2013
|
|
4A7G
 
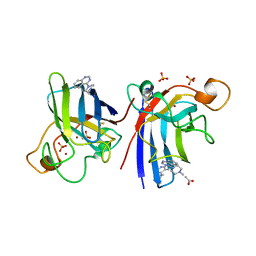 | | Structure of human I113T SOD1 mutant complexed with 4-methylpiperazin- 1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)QUINAZOLINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Sharma, R, Antonyuk, S.V, Strange, R.W, Berry, N.G, O'Neil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Curr.Med.Chem., 20, 2013
|
|
6T4B
 
 | | CRYSTAL STRUCTURE OF HUMAN TDP-43 N-TERMINAL DOMAIN AT 2.55 A RESOLUTION | | Descriptor: | SULFATE ION, TAR DNA-binding protein 43 | | Authors: | Watanabe, T.F, Wright, G.S.A, Amporndanai, K, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-10-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Purification and Structural Characterization of Aggregation-Prone Human TDP-43 Involved in Neurodegenerative Diseases.
Iscience, 23, 2020
|
|
6THF
 
 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
4B3E
 
 | | Structure of copper-zinc superoxide dismutase complexed with bicarbonate. | | Descriptor: | CARBONATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Hough, M.A, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Evidence for a Copper-Bound Carbonate Intermediate in the Peroxidase and Dismutase Activities of Superoxide Dismutase.
Plos One, 7, 2012
|
|
4A7V
 
 | | Structure of human I113T SOD1 mutant complexed with dopamine in the p21 space group | | Descriptor: | COPPER (II) ION, L-DOPAMINE, SULFATE ION, ... | | Authors: | Wright, G.S.A, Antonyuk, S.V, Kershaw, N.M, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
6T6V
 
 | | Glu-494-Ala inactive monomer of a quinol dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, Nitric oxide reductase subunit B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gopalasingam, C.C, Johnson, R.M, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-10-19 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
4CSZ
 
 | | STRUCTURE OF F306C MUTANT OF NITRITE REDUCTASE FROM Achromobacter XYLOSOXIDANS WITH NITRITE BOUND | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Leferink, N.G.H, Antonyuk, S.V, Houwman, J.A, Scrutton, N.S, REady, R, Hasnain, S.S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of Residues Remote from the Catalytic Centre on Enzyme Catalysis of Copper Nitrite Reductase.
Nat.Commun., 5, 2014
|
|
4A7S
 
 | | Structure of human I113T SOD1 mutant complexed with 5-Fluorouridine in the p21 space group | | Descriptor: | 5-FLUOROURIDINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-12-05 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
5NVD
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria at 2.5 A resolution in P6322 crystal form | | Descriptor: | CBS-CP12 | | Authors: | Hackenberg, C, Hakanpaa, J, Eigner, C, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6TFD
 
 | | Crystal structure of nitrite and NO bound three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
6SW6
 
 | |
4A7U
 
 | | Structure of human I113T SOD1 complexed with adrenaline in the p21 space group. | | Descriptor: | ACETATE ION, COPPER (II) ION, L-EPINEPHRINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4D6T
 
 | | Cytochrome bc1 bound to the 4(1H)-pyridone GW844520 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-chloro-2,6-dimethyl-5-{4-[4-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, CARDIOLIPIN, ... | | Authors: | Capper, M.J, O'Neill, P.M, Fisher, N, Strange, R.W, Moss, D, Ward, S.A, Berry, N.G, Lawrenson, A.S, Hasnain, S.S, Biagini, G.A, Antonyuk, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Antimalarial 4(1H)-Pyridones Bind to the Qi Site of Cytochrome Bc1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4D6U
 
 | | Cytochrome bc1 bound to the 4(1H)-pyridone GSK932121 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, CARDIOLIPIN, ... | | Authors: | Capper, M.J, ONeill, P.M, Fisher, N, Strange, R.W, Moss, D, Ward, S.A, Berry, N.G, Lawrenson, A.S, Hasnain, S.S, Biagini, G.A, Antonyuk, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.09 Å) | | Cite: | Antimalarial 4(1H)-Pyridones Bind to the Qi Site of Cytochrome Bc1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6THE
 
 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6TFO
 
 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
2VM4
 
 | | Structure of Alcaligenes xylosoxidans nitrite reductase in space group R3 - 2 of 2 | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallography with Online Optical and X-Ray Absorption Spectroscopies Demonstrates an Ordered Mechanism in Copper Nitrite Reductase.
J.Mol.Biol., 378, 2008
|
|
2V8U
 
 | | Atomic resolution structure of Mn catalase from Thermus Thermophilus | | Descriptor: | LITHIUM ION, MANGANESE (II) ION, MANGANESE-CONTAINING PSEUDOCATALASE, ... | | Authors: | Barynin, V.V, Antonyuk, S.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
2VM3
 
 | | Structure of Alcaligenes xylosoxidans in space group R3 - 1 of 2 | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallography with Online Optical and X-Ray Absorption Spectroscopies Demonstrates an Ordered Mechanism in Copper Nitrite Reductase.
J.Mol.Biol., 378, 2008
|
|
5M6C
 
 | | CRYSTAL STRUCTURE OF T71N MUTANT OF HUMAN HIPPOCALCIN | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Helassa, N, Antonyuk, S.V, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5NMI
 
 | | Cytochrome bc1 bound to the inhibitor MJM170 | | Descriptor: | (4aS)-2-methyl-3-(4-phenoxyphenyl)-5,6,7,8-tetrahydroquinolin-4(4aH)-one, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ARG-ASN-TRP-VAL-PRO-THR-ALA-GLN-LEU-TRP-GLY-ALA-VAL-GLY-ALA-VAL-GLY-LEU-VAL-SER-ALA-THR, ... | | Authors: | Capper, N.J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-14 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New paradigms for understanding and step changes in treating active and chronic, persistent apicomplexan infections.
Sci Rep, 6, 2016
|
|
