1R4L
 
 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1RIH
 
 | | Crystal Structure of Fab 14F7, a unique anti-tumor antibody specific for N-glycolyl GM3 | | Descriptor: | heavy chain of antibody 14F7, light chain of antibody 14F7 | | Authors: | Krengel, U, Olsson, L.-L, Martinez, C, Talavera, A, Rojas, G, Mier, E, Angstrom, J, Moreno, E. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Molecular Interactions of a Unique Antitumor Antibody Specific for N-Glycolyl GM3.
J.Biol.Chem., 279, 2004
|
|
1NR7
 
 | | Crystal structure of apo bovine glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1M1C
 
 | | Structure of the L-A virus | | Descriptor: | Major coat protein | | Authors: | Naitow, H, Tang, J, Canady, M, Wickner, R.B, Johnson, J.E. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism.
Nat.Struct.Biol., 9, 2002
|
|
1N4M
 
 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
1S28
 
 | | Crystal Structure of AvrPphF ORF1, the Chaperone for the Type III Effector AvrPphF ORF2 from P. syringae | | Descriptor: | ORF1, SULFATE ION | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.L, Sondek, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
1WT9
 
 | | crystal structure of Aa-X-bp-I, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, agkisacutacin A chain, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-18 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
1O5D
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, Coagulation factor VII, Tissue factor | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1OLS
 
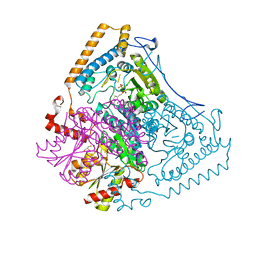 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1ITZ
 
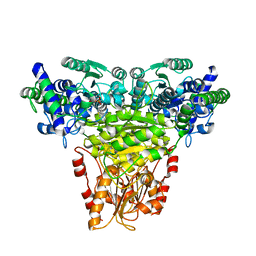 | | Maize Transketolase in complex with TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Gerhardt, S, Echt, S, Bader, G, Freigang, J, Busch, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-15 | | Release date: | 2003-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and properties of an engineered transketolase from maize
PLANT PHYSIOL., 132, 2003
|
|
1SZU
 
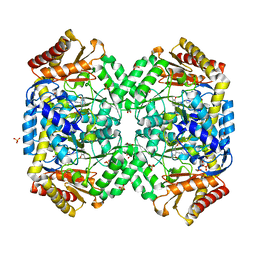 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1O5A
 
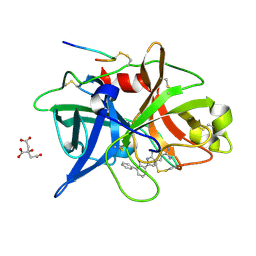 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1N6A
 
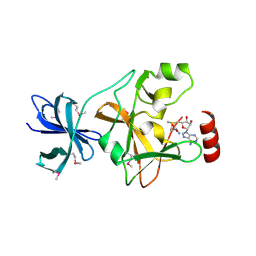 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6C
 
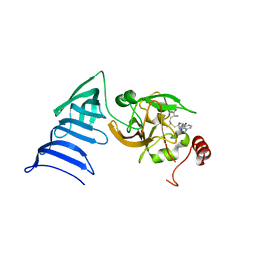 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6V
 
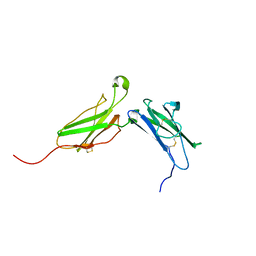 | | Average structure of the interferon-binding ectodomain of the human type I interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
1MKY
 
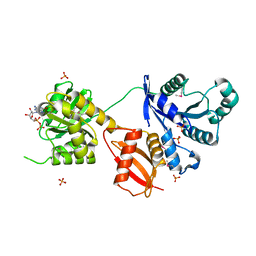 | | Structural Analysis of the Domain Interactions in Der, a Switch Protein Containing Two GTPase Domains | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Probable GTP-binding protein engA | | Authors: | Robinson, V.L, Hwang, J, Fox, E, Inouye, M, Stock, A.M. | | Deposit date: | 2002-08-29 | | Release date: | 2003-01-14 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain Arrangement of Der, a Switch Protein Containing Two GTPase Domains
Structure, 10, 2002
|
|
1R42
 
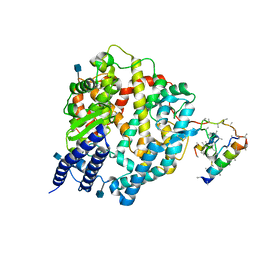 | | Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ZINC ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1BXP
 
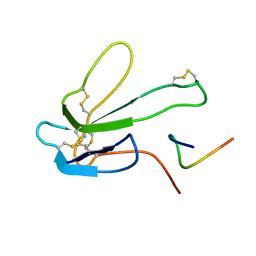 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, 20 STRUCTURES | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE MET-ARG-TYR-TYR-GLU-SER-SER-LEU-LYS-SER-TYR-PRO-ASP | | Authors: | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | Deposit date: | 1998-08-23 | | Release date: | 1999-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1U6U
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
1O5C
 
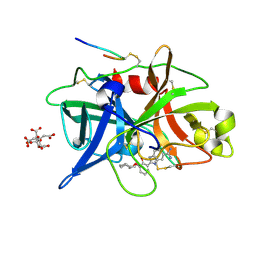 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1NQT
 
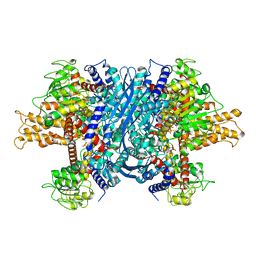 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1O5B
 
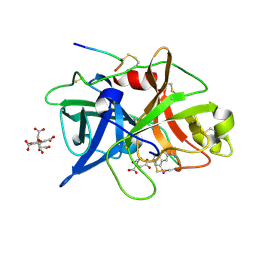 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5G
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CALCIUM ION, Hirudin IIIB', ... | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1S21
 
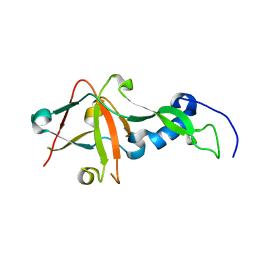 | | Crystal Structure of AvrPphF ORF2, A Type III Effector from P. syringae | | Descriptor: | ORF2 | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.K, Sondek, J. | | Deposit date: | 2004-01-07 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
1SF2
 
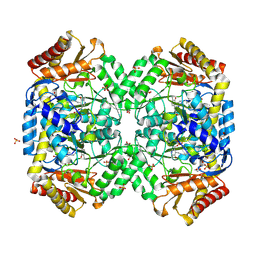 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
