7KKI
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 2,4-Dichlorophenoxyacetic acid | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KIG
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole-3-butyric acid | | Descriptor: | 3-INDOLEBUTYRIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KYM
 
 | |
2BAZ
 
 | |
1NMI
 
 | | Solution structure of the imidazole complex of iso-1 cytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C, ... | | Authors: | Yao, Y, Tong, Y, Liu, G, Wang, J, Zheng, J, Tang, W. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the imidazole complex of iso-1 cytochrome c
To be Published
|
|
1AXA
 
 | | ACTIVE-SITE MOBILITY IN HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE AS DEMONSTRATED BY CRYSTAL STRUCTURE OF A28S MUTANT | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Hartsuck, J.A, Foundling, S, Ermolieff, J, Tang, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant.
Protein Sci., 7, 1998
|
|
2CEU
 
 | | Despentapeptide insulin in acetic acid (pH 2) | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Zhang, Y, Zakova, L, Dodson, E.J, Turkenburg, J.P, Brange, J, Dodson, G.G. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | I222 Crystal Form of Despentapeptide (B26-B30) Insulin Provides New Insights Into the Properties of Monomeric Insulin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3EFX
 
 | | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin, 1.9A crystal structure reveals the details | | Descriptor: | Cholera enterotoxin subunit B, Heat-labile enterotoxin B chain, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose | | Authors: | Holmner, A, Lebens, M, Teneberg, S, Angstrom, J, Okvist, M, Krengel, U. | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin: 1.9 A crystal structure reveals the details
Structure, 12, 2004
|
|
2ZU6
 
 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
2Q8I
 
 | | Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, GLYCEROL, ... | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
1NY8
 
 | | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115 | | Descriptor: | Protein yrbA | | Authors: | Swapna, G.V.T, Huang, J.Y, Acton, T.B, Shastry, R, Chiang, Y.-W, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115
To be Published
|
|
2V5Q
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE PLK-1 KINASE DOMAIN IN COMPLEX WITH A SELECTIVE DARPIN | | Descriptor: | DESIGN ANKYRIN REPEAT PROTEIN, SERINE/THREONINE-PROTEIN KINASE PLK1 | | Authors: | Bandeiras, T.M, Hillig, R.C, Matias, P.M, Eberspaecher, U, Fanghaenel, J, Thomaz, M, Miranda, S, Crusius, K, Puetter, V, Amstutz, P, Gulotti-Georgieva, M, Binz, H.K, Holz, C, Schmitz, A.A.P, Lang, C, Donner, P, Egner, U, Carrondo, M.A, Mueller-Tiemann, B. | | Deposit date: | 2007-07-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of wild-type Plk-1 kinase domain in complex with a selective DARPin.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
2F60
 
 | | Crystal Structure of the Dihydrolipoamide Dehydrogenase (E3)-Binding Domain of Human E3-Binding Protein | | Descriptor: | GLYCEROL, Pyruvate dehydrogenase protein X component | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
3EAM
 
 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
1FKN
 
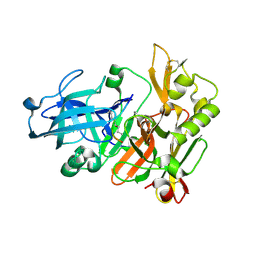 | | Structure of Beta-Secretase Complexed with Inhibitor | | Descriptor: | MEMAPSIN 2, inhibitor | | Authors: | Hong, L, Koelsch, G, Lin, X, Wu, S, Terzyan, S, Ghosh, A, Zhang, X.C, Tang, J. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor.
Science, 290, 2000
|
|
2OCF
 
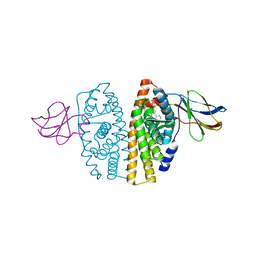 | | Human estrogen receptor alpha ligand-binding domain in complex with estradiol and the E2#23 FN3 monobody | | Descriptor: | ESTRADIOL, Estrogen receptor, Fibronectin | | Authors: | Rajan, S.S, Kuruvilla, S.M, Sharma, S.K, Kim, Y, Huang, J, Koide, A, Koide, S, Joachimiak, A, Greene, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2KND
 
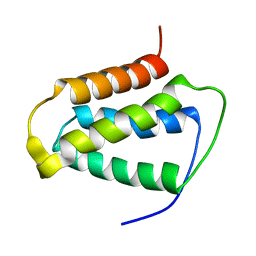 | | Psb27 structure from Synechocystis | | Descriptor: | Photosystem II 11 kDa protein | | Authors: | Cormann, K.U, Nowaczyk, M.M, Bangert, J.-A, Ikeuchi, M, Roegner, M, Stoll, R. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Psb27 in solution: implications for transient binding to photosystem II during biogenesis and repair
Biochemistry, 48, 2009
|
|
1BGW
 
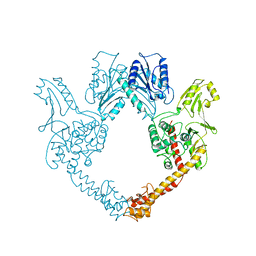 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
1FXQ
 
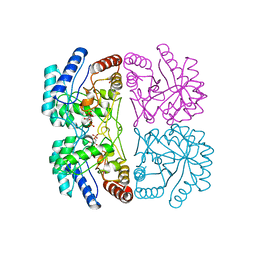 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND A5P | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-26 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
1FWT
 
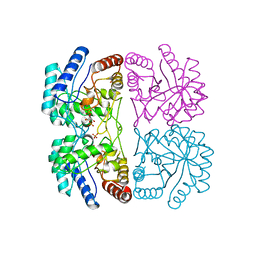 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP, E4P AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, ERYTHOSE-4-PHOSPHATE, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
1U6B
 
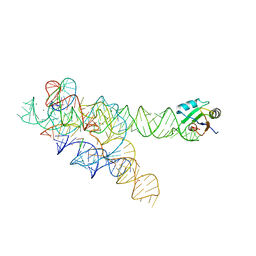 | | CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS | | Descriptor: | 197-MER, 5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*AP*GP*AP*CP*GP *GP*CP*C)-3', 5'-R(*CP*AP*(5MU))-3', ... | | Authors: | Adams, P.L, Stahley, M.R, Kosek, A.B, Wang, J, Strobel, S.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Self-Splicing Group I Intron with Both Exons.
Nature, 430, 2004
|
|
1UAJ
 
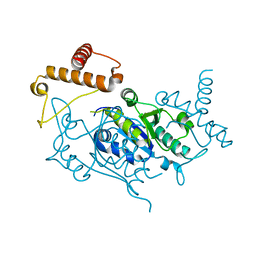 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1C2A
 
 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1UAM
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
