6KRW
 
 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | 分子名称: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | 著者 | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | 登録日 | 2019-08-22 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.24916625 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
4XX0
 
 | | CoA bound to pig GTP-specific succinyl-CoA synthetase | | 分子名称: | COENZYME A, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Fraser, M.E, Huang, J, Malhi, M. | | 登録日 | 2015-01-29 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of GTP-specific succinyl-CoA synthetase in complex with CoA.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6KYV
 
 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | 分子名称: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | 著者 | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | 登録日 | 2019-09-20 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
6ASY
 
 | | BiP-ATP2 | | 分子名称: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | 著者 | Liu, Q, Yang, J, Zong, Y, Columbus, L, Zhou, L. | | 登録日 | 2017-08-26 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s.
Nat Commun, 8, 2017
|
|
3PQ1
 
 | | Crystal structure of human mitochondrial poly(A) polymerase (PAPD1) | | 分子名称: | Poly(A) RNA polymerase | | 著者 | Bai, Y, Srivastava, S.K, Chang, J.H, Tong, L. | | 登録日 | 2010-11-25 | | 公開日 | 2011-03-30 | | 最終更新日 | 2017-08-02 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase.
Mol.Cell, 41, 2011
|
|
6L0Y
 
 | | Structure of dsRNA with G-U wobble base pairs | | 分子名称: | RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3') | | 著者 | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of dsRNA with G-U wobble base pairs
To Be Published
|
|
3PXG
 
 | | Structure of MecA121 and ClpC1-485 complex | | 分子名称: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-09 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.654 Å) | | 主引用文献 | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
3PZ7
 
 | |
8KCA
 
 | |
3PS4
 
 | | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1 | | 分子名称: | 1,2-ETHANEDIOL, IMIDAZOLE, Microtubule-associated serine/threonine-protein kinase 1 | | 著者 | Ugochukwu, E, Wang, J, Krojer, T, Muniz, J.R.C, Sethi, R, Pike, A.C.W, Roos, A, Salah, E, Cocking, R, Savitsky, P, Doyle, D.A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Elkins, J.M, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-30 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1
TO BE PUBLISHED
|
|
5GMQ
 
 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | 分子名称: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | 著者 | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | 登録日 | 2016-07-15 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
4E3R
 
 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | 分子名称: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | 著者 | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | 登録日 | 2012-03-10 | | 公開日 | 2012-10-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
8SSQ
 
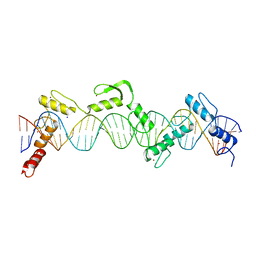 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | 分子名称: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
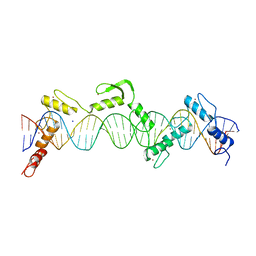 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | 分子名称: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
6KQW
 
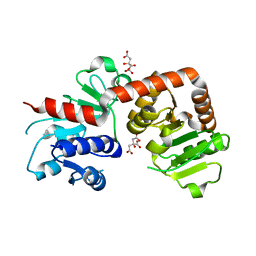 | | Crystal structure of Yijc from B. subtilis | | 分子名称: | CITRIC ACID, Uncharacterized UDP-glucosyltransferase YjiC | | 著者 | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
3FWE
 
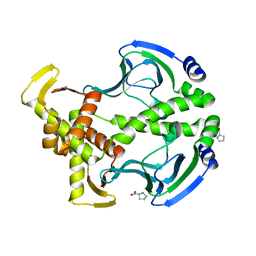 | | Crystal Structure of the Apo D138L CAP mutant | | 分子名称: | Catabolite gene activator, PROLINE | | 著者 | Sharma, H, Wang, J, Kong, J, Yu, S, Steitz, T. | | 登録日 | 2009-01-17 | | 公開日 | 2009-09-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8S9B
 
 | | Cryo-EM structure of Nav1.7 with LCM | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fan, X, Huang, J, Yan, N. | | 登録日 | 2023-03-27 | | 公開日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
6KWT
 
 | |
8S9C
 
 | | Cryo-EM structure of Nav1.7 with CBZ | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fan, X, Huang, J, Yan, N. | | 登録日 | 2023-03-27 | | 公開日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | 分子名称: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | 著者 | Hare, S, Wang, J, Cherepanov, P. | | 登録日 | 2009-06-04 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
5UIS
 
 | | Crystal structure of IRAK4 in complex with compound 12 | | 分子名称: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | 著者 | Han, S, Chang, J.S. | | 登録日 | 2017-01-14 | | 公開日 | 2017-05-24 | | 最終更新日 | 2017-07-26 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
6L8W
 
 | | Crystal structure of ugt transferase mutant2 | | 分子名称: | Glycosyltransferase | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2019-11-07 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
5GPH
 
 | |
2M2R
 
 | | Solution structure of MCh-2: A novel inhibitor cystine knot peptide from Momordica charantia | | 分子名称: | Inhibitor cystine knot peptide MCh-2 | | 著者 | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | 登録日 | 2013-01-01 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
