5UN8
 
 | |
2KQ3
 
 | | Solution structure of SNase140 | | Descriptor: | Thermonuclease | | Authors: | Wang, M, Feng, Y, Yao, H, Wang, J. | | Deposit date: | 2009-10-26 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Importance of the C-Terminal Loop L137-S141 for the Folding and Folding Stability of Staphylococcal Nuclease
Biochemistry, 49, 2010
|
|
1CJY
 
 | | HUMAN CYTOSOLIC PHOSPHOLIPASE A2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, PROTEIN (CYTOSOLIC PHOSPHOLIPASE A2) | | Authors: | Dessen, A, Tang, J, Schmidt, H, Stahl, M, Clark, J.D, Seehra, J, Somers, W.S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
4NK7
 
 | |
2F60
 
 | | Crystal Structure of the Dihydrolipoamide Dehydrogenase (E3)-Binding Domain of Human E3-Binding Protein | | Descriptor: | GLYCEROL, Pyruvate dehydrogenase protein X component | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
7L9A
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2KQE
 
 | | Second PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KX6
 
 | | Signaling state of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Ramachandran, P.L, Lovett, J.E, Carl, P.J, Cammarata, M, Lee, J.H, Yang, J.O, Ihee, H, Timmel, C.R, van Thor, J. | | Deposit date: | 2010-04-27 | | Release date: | 2011-06-15 | | Last modified: | 2012-07-18 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The short-lived signaling state of the photoactive yellow protein photoreceptor revealed by combined structural probes.
J.Am.Chem.Soc., 133, 2011
|
|
2KU3
 
 | |
2KRL
 
 | | The ensemble of the solution global structures of the 102-nt ribosome binding structure element of the turnip crinkle virus 3' UTR RNA | | Descriptor: | RNA (102-MER) | | Authors: | Zuo, X, Wang, J, Yu, P, Eyler, D, Xu, H, Starich, M, Tiede, D, Simon, A, Kasprzak, W, Schwieters, C, Shapiro, B. | | Deposit date: | 2009-12-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5UN9
 
 | | The crystal structure of human O-GlcNAcase in complex with Thiamet-G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J. | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2F76
 
 | | Solution structure of the M-PMV wild type matrix protein (p10) | | Descriptor: | Core protein p10 | | Authors: | Vlach, J, Lipov, J, Veverka, V, Lang, J, Srb, P, Rumlova, M, Hunter, E, Ruml, T, Hrabal, R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2KQT
 
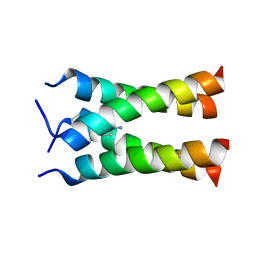 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
2L3T
 
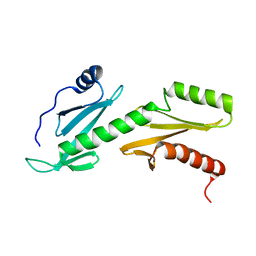 | | Solution structure of tandem SH2 domain from Spt6 | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Liu, J, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-09-22 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain
To be Published
|
|
2LA9
 
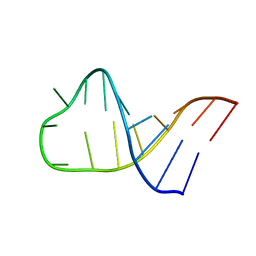 | | NMR structure of Pseudouridine_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*(PSU)P*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
3CD8
 
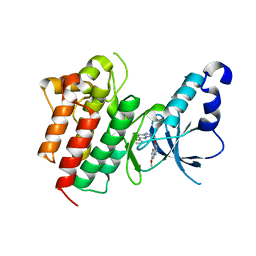 | | X-ray Structure of c-Met with triazolopyridazine Inhibitor. | | Descriptor: | 7-methoxy-4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methoxy]quinoline, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Albrecht, B.K, Harmange, J.-C, Bauer, D, Choquette, D, Dussault, I. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
1HF9
 
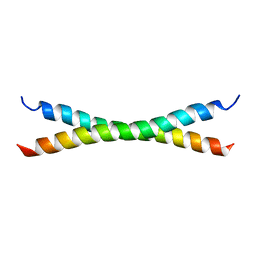 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
2L43
 
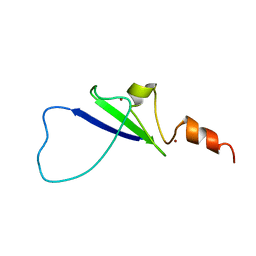 | | Structural basis for histone code recognition by BRPF2-PHD1 finger | | Descriptor: | Histone H3.3,LINKER,Bromodomain-containing protein 1, ZINC ION | | Authors: | Qin, S, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of unmodified histone H3 by the first PHD finger of Bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases MOZ and MORF
To be Published
|
|
2F5G
 
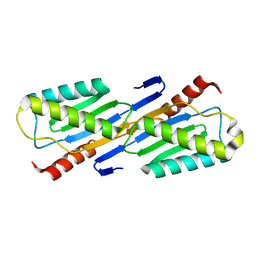 | | Crystal structure of IS200 transposase | | Descriptor: | Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2F5Z
 
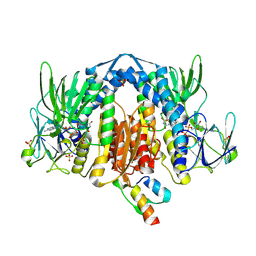 | | Crystal Structure of Human Dihydrolipoamide Dehydrogenase (E3) Complexed to the E3-Binding Domain of Human E3-Binding Protein | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate dehydrogenase protein X component, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
2F4F
 
 | | Crystal structure of IS200 transposase | | Descriptor: | MANGANESE (II) ION, Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2JYH
 
 | |
2L7B
 
 | | NMR Structure of full length apoE3 | | Descriptor: | Apolipoprotein E | | Authors: | Chen, J, Wang, J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LEO
 
 | |
2L7E
 
 | |
