4JCU
 
 | | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1 | | Descriptor: | 5-carboxymethyl-2-hydroxymuconate isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1
To be Published
|
|
1S0G
 
 | |
1TXZ
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme.
Protein Sci., 14, 2005
|
|
3U4J
 
 | | Crystal structure of NAD-dependent aldehyde dehydrogenase from Sinorhizobium meliloti | | Descriptor: | CALCIUM ION, NAD-dependent aldehyde dehydrogenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NAD-dependent aldehyde dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3U4F
 
 | | Crystal structure of a mandelate racemase (muconate lactonizing enzyme family protein) from Roseovarius nubinhibens | | Descriptor: | GUANIDINE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Eswaramoorthy, S, Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a mandelate racemase (muconate lactonizing enzyme family protein) from Roseovarius nubinhibens
To be Published, 2011
|
|
1TY8
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YMX7
To be Published
|
|
1TYH
 
 | |
4JWK
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Acinetobacter baumannii with cytidine at 1.87 A resolution | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
1S0E
 
 | | Crystal structure of botulinum neurotoxin type B at pH 6.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1RTT
 
 | | Crystal structure determination of a putative NADH-dependent reductase using sulfur anomalous signal | | Descriptor: | SULFATE ION, conserved hypothetical protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2G6T
 
 | |
1ZKX
 
 | | Crystal structure of Glu158Ala/Thr159Ala/Asn160Ala- a triple mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
3UP8
 
 | | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B | | Descriptor: | ACETATE ION, Putative 2,5-diketo-D-gluconic acid reductase B | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B
To be Published
|
|
1Y9Q
 
 | |
1YDF
 
 | |
4LK5
 
 | | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-06 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10
To be Published
|
|
1RV9
 
 | |
1RRM
 
 | |
5DA3
 
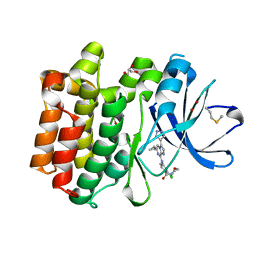 | | Crystal structure of PTK6 Kinase domain with inhibitor | | Descriptor: | (2-chloro-4-{[6-cyclopropyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}phenyl)(morpholin-4-yl)methanone, GLYCEROL, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Battula, S.K, Vadivelu, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
4JFC
 
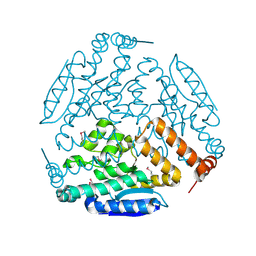 | | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666 | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666
To be Published
|
|
1S0B
 
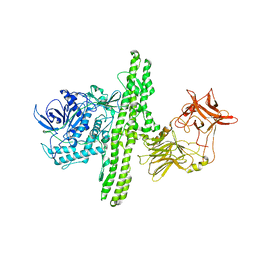 | | Crystal structure of botulinum neurotoxin type B at pH 4.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
3NND
 
 | |
1ZL6
 
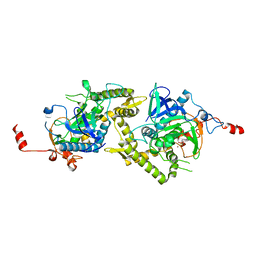 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1ZN3
 
 | | Crystal structure of Glu335Ala mutant of Clostridium botulinum neurotoxin type E | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-11 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
4KPK
 
 | | Crystal structure of a enoyl-CoA hydratase from Shewanella pealeana ATCC 700345 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-13 | | Release date: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Shewanella pealeana ATCC 700345
To be Published
|
|
