1TEL
 
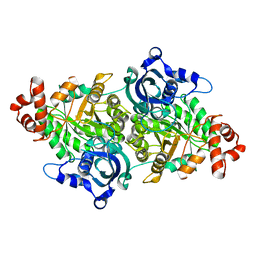 | | Crystal structure of a RubisCO-like protein from Chlorobium tepidum | | Descriptor: | ribulose bisphosphate carboxylase, large subunit | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-05-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a RubisCO-like protein from Chlorobium tepidum
To be Published
|
|
1TE5
 
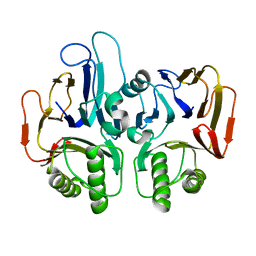 | |
1T5O
 
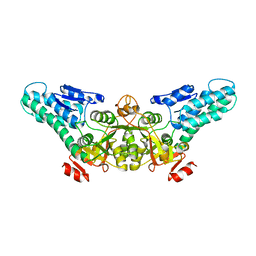 | | Crystal structure of the translation initiation factor eIF-2B, subunit delta, from A. fulgidus | | Descriptor: | Translation initiation factor eIF2B, subunit delta | | Authors: | Malashkevich, V.N, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the translation initiation factor eIF-2B, subunit delta, from
A. fulgidus
to be published
|
|
1T62
 
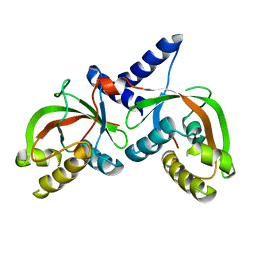 | |
1WHO
 
 | | ALLERGEN PHL P 2 | | Descriptor: | ALLERGEN PHL P 2 | | Authors: | Fedorov, A.A, Fedorov, E.V, Dolecek, C, Susani, M, Valenta, R, Almo, S.C. | | Deposit date: | 1997-04-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Phl P 2, a Major Timothy Grass (Phleum Pratense) Pollen Allergen
To be Published
|
|
1SV6
 
 | | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli | | Descriptor: | 2-keto-4-pentenoate hydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sharp, A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-03-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli
To be Published
|
|
1WVI
 
 | |
1SVY
 
 | | SEVERIN DOMAIN 2, 1.75 ANGSTROM CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, SEVERIN, SODIUM ION | | Authors: | Puius, Y.A, Fedorov, E.V, Eichinger, L, Sullivan, M, Schleicher, M, Almo, S.C. | | Deposit date: | 1998-08-10 | | Release date: | 1999-08-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the functional surface of domain 2 in the gelsolin superfamily.
Biochemistry, 39, 2000
|
|
1WV2
 
 | |
1WUE
 
 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
1JUF
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
1K27
 
 | | Crystal Structure of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase in Complex with a Transition State Analogue | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-Deoxy-5'-Methylthioadenosine Phosphorylase, PHOSPHATE ION | | Authors: | Shi, W, Singh, V, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-09-26 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Picomolar transition state analogue inhibitors of human 5'-methylthioadenosine phosphorylase and X-ray structure with MT-immucillin-A
Biochemistry, 43, 2004
|
|
1K4Z
 
 | | C-terminal Domain of Cyclase Associated Protein | | Descriptor: | Adenylyl Cyclase-Associated Protein | | Authors: | Rozwarski, D.A, Fedorov, A.A, Dodatko, T, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-09 | | Release date: | 2002-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein
Biochemistry, 43, 2004
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
1LH0
 
 | | Crystal Structure of Salmonella typhimurium OMP Synthase in Complex with MGPRPP and Orotate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, OMP synthase, ... | | Authors: | Fedorov, A.A, Panneerselvam, K, Shi, W, Grubmeyer, C, Almo, S.C. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Salmonella typhimurium OMP Synthase in a Complete Substrate Complex.
Biochemistry, 51, 2012
|
|
1L1R
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
1LCU
 
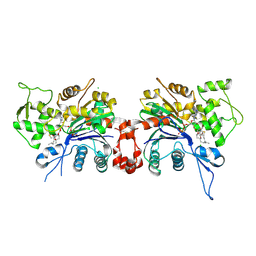 | | Polylysine Induces an Antiparallel Actin Dimer that Nucleates Filament Assembly: Crystal Structure at 3.5 A Resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Bubb, M.R, Govindasamy, L, Yarmola, E.G, Vorobiev, S.M, Almo, S.C, Somasundaram, T, Chapman, M.S, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2002-04-06 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Polylysine induces an antiparallel actin dimer that nucleates filament assembly: crystal structure at 3.5-A resolution.
J.Biol.Chem., 277, 2002
|
|
1L9G
 
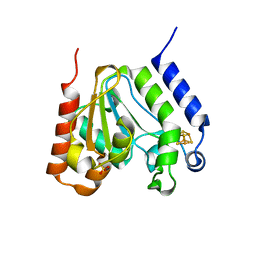 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1QPV
 
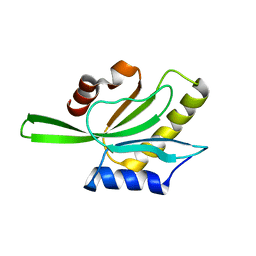 | | YEAST COFILIN | | Descriptor: | YEAST COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1999-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1M9P
 
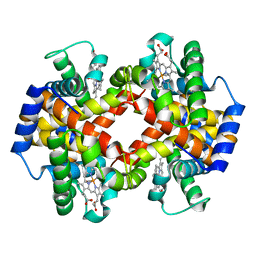 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5ETJ
 
 | | Crystal structure of purine nucleoside phosphorylase (E258D, L261A) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Suarez, J, Schramm, V.L, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulating Enzyme Catalysis through Mutations Designed to Alter Rapid Protein Dynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5EUB
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 2-amino-MTA and sulfate | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-[2,6-bis(azanyl)purin-9-yl]-5-(methylsulfanylmethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 2-amino-MTA and sulfate
To be published
|
|
5EPE
 
 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
