8FYG
 
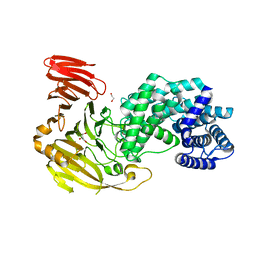 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | 著者 | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | 登録日 | 2023-01-26 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8FNX
 
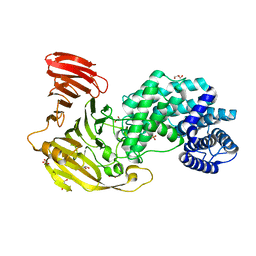 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | 分子名称: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | 著者 | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | 登録日 | 2022-12-28 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
5WW0
 
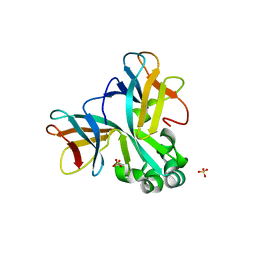 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | 分子名称: | SET domain-containing protein 7, SULFATE ION | | 著者 | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | 登録日 | 2016-12-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
8G0O
 
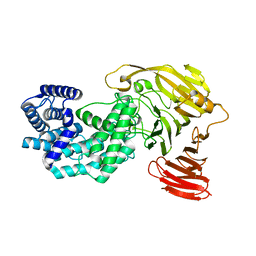 | | Crystal structure of Y281F mutant of Hyaluronate lyase B from Cutibacterium acnes | | 分子名称: | Hyaluronate lyase | | 著者 | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
4GDE
 
 | |
7ZLG
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | 分子名称: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | 著者 | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | 登録日 | 2022-04-15 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-05-10 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | 分子名称: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | 著者 | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | 登録日 | 2022-04-15 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-05-10 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
8AGB
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide bound | | 分子名称: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | 登録日 | 2022-07-19 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
8AGC
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide and non-acceptor peptide bound | | 分子名称: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | 登録日 | 2022-07-19 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
7ZYI
 
 | | Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody | | 分子名称: | CHOLESTEROL, GLYCOCHENODEOXYCHOLIC ACID, Nanobody, ... | | 著者 | Liu, H, Irobalieva, R.N, Bang-Sorensen, R, Nosol, K, Mukherjee, S, Agrawal, P, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | 登録日 | 2022-05-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver.
Cell Res., 32, 2022
|
|
7ZLH
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | | 分子名称: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | 著者 | Bloch, J.S, Mukherjee, S, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | 登録日 | 2022-04-15 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-05-10 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
2Y3M
 
 | | Structure of the extra-membranous domain of the secretin HofQ from Actinobacillus actinomycetemcomitans | | 分子名称: | FORMIC ACID, GLYCEROL, PROTEIN TRANSPORT PROTEIN HOFQ | | 著者 | Tarry, M, Jaaskelainen, M, Tuominen, H, Ihalin, R, Hogbom, M. | | 登録日 | 2010-12-21 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Extra-Membranous Domains of the Competence Protein Hofq Show DNA Binding, Flexibility and a Shared Fold with Type I Kh Domains.
J.Mol.Biol., 409, 2011
|
|
4GJH
 
 | | Crystal Structure of the TRAF domain of TRAF5 | | 分子名称: | TNF receptor-associated factor 5 | | 著者 | Zhang, P, Reichardt, A, Liang, H, Wang, Y, Cheng, D, Aliyari, R, Cheng, G, Liu, Y. | | 登録日 | 2012-08-09 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5
Sci.Signal., 5, 2012
|
|
7OCI
 
 | | Cryo-EM structure of yeast Ost6p containing oligosaccharyltransferase complex | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wild, R, Neuhaus, J.D, Eyring, J, Irobalieva, R.N, Kowal, J, Lin, C.W, Locher, K.P, Aebi, M. | | 登録日 | 2021-04-27 | | 公開日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Functional analysis of Ost3p and Ost6p containing yeast oligosaccharyltransferases.
Glycobiology, 31, 2021
|
|
7BB1
 
 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Glutamic acid 2:1 | | 分子名称: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | 著者 | Belviso, B.D, Caliandro, R, Carrozzini, B. | | 登録日 | 2020-12-16 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
7B9J
 
 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Urea 1:2 | | 分子名称: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | 著者 | Belviso, B.D, Caliandro, R, Carrozzini, B. | | 登録日 | 2020-12-14 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
7BAZ
 
 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Glycerol 1:2 | | 分子名称: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | 著者 | Belviso, B.D, Caliandro, R, Carrozzini, B. | | 登録日 | 2020-12-16 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
7MU9
 
 | |
8DPK
 
 | |
8STH
 
 | | human STING with diABZI agonist 15 | | 分子名称: | 1-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | 著者 | Duvall, J.R, Bukhalid, R.A. | | 登録日 | 2023-05-10 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
7ZC3
 
 | | Crystal structure of human copper chaperone Atox1 bound to zinc ion by CxxC motif | | 分子名称: | Copper transport protein ATOX1, SULFATE ION, ZINC ION | | 著者 | Mangini, V, Belviso, B.D, Arnesano, F, Caliandro, R. | | 登録日 | 2022-03-25 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Human Copper Chaperone ATOX1 Bound to Zinc Ion.
Biomolecules, 12, 2022
|
|
1TC1
 
 | | A 1.4 ANGSTROM CRYSTAL STRUCTURE FOR THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE OF TRYPANOSOMA CRUZI | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMYCIN B, PROTEIN (HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE) | | 著者 | Focia, P.J, Craig III, S.P, Nieves-Alicea, R, Fletterick, R.J, Eakin, A.E. | | 登録日 | 1998-09-30 | | 公開日 | 1999-10-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
Biochemistry, 37, 1998
|
|
8STI
 
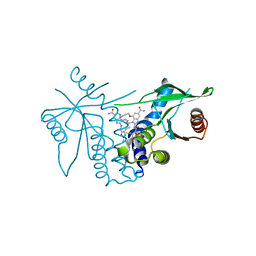 | | human STING with agonist XMT-1616 | | 分子名称: | 3-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-3H-imidazo[4,5-b]pyridine-6-carboxamide, Stimulator of interferon genes protein | | 著者 | Duvall, J.R, Bukhalid, R.A. | | 登録日 | 2023-05-10 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
4P7E
 
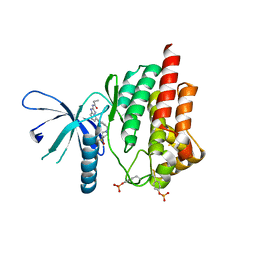 | | Triazolopyridine compounds as selective JAK1 inhibitors: from hit identification to GLPG0634 | | 分子名称: | N-(5-{4-[(1,1-dioxidothiomorpholin-4-yl)methyl]phenyl}[1,2,4]triazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide, Tyrosine-protein kinase JAK2 | | 著者 | Menet, C.C.J, Fletcher, S, Van Lommen, G, Geney, R, Blanc, J, Smits, K, Jouannigot, N, van der Aar, E.M, Clement-Lacroix, P, Lepescheux, L, Galien, R, Vayssiere, B, Nelles, L, Christophe, T, Brys, R, Uhring, M, Ciesielski, F, Van Rompaey, L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Triazolopyridines as Selective JAK1 Inhibitors: From Hit Identification to GLPG0634.
J.Med.Chem., 57, 2014
|
|
1L1M
 
 | | SOLUTION STRUCTURE OF A DIMER OF LAC REPRESSOR DNA-BINDING DOMAIN COMPLEXED TO ITS NATURAL OPERATOR O1 | | 分子名称: | 5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3', Lactose operon repressor | | 著者 | Kalodimos, C.G, Bonvin, A.M.J.J, Salinas, R.K, Wechselberger, R, Boelens, R, Kaptein, R. | | 登録日 | 2002-02-19 | | 公開日 | 2002-06-26 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain.
EMBO J., 21, 2002
|
|
