6O44
 
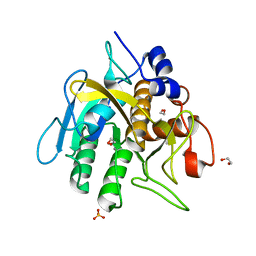 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-02-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
8FR5
 
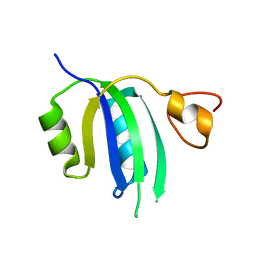 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
9CLR
 
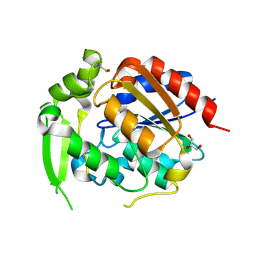 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seventy-one Mutations | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tan, P, Meixner, E.L, Nguyen, A, Kazlaukas, R.J, Pierce, C.T, Evans, R.L, Shi, K, Aihara, H. | | Deposit date: | 2024-07-12 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Hydroxynitrile Lyase from Hevea brasiliensis with Seventy-one Mutations
To Be Published
|
|
1Z1B
 
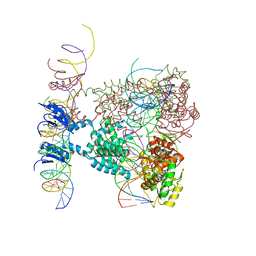 | | Crystal structure of a lambda integrase dimer bound to a COC' core site | | Descriptor: | 26-MER DNA, 29-MER DNA, 5'-D(*CP*T*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6P7A
 
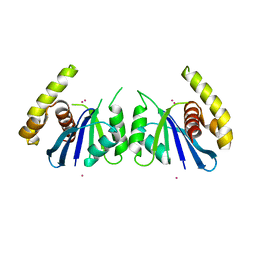 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | Descriptor: | CADMIUM ION, Holliday junction resolvase | | Authors: | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
6DRT
 
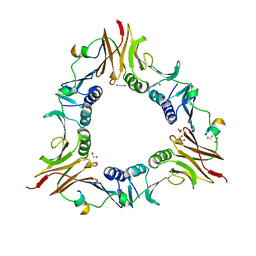 | |
6NFK
 
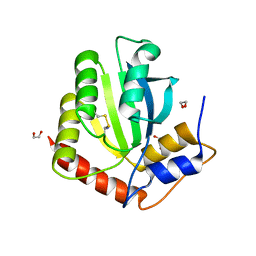 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NFM
 
 | |
5U0L
 
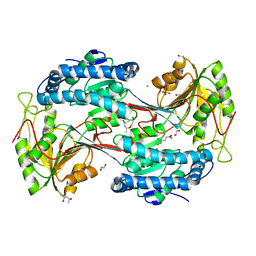 | | X-ray crystal structure of fatty aldehyde dehydrogenase enzymes from Marinobacter aquaeolei VT8 complexed with a substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
6P7B
 
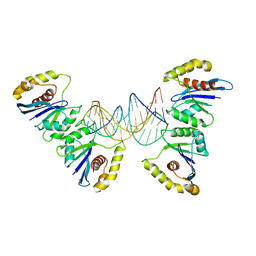 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | Descriptor: | DNA (29-MER), Holliday junction resolvase | | Authors: | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
5CQK
 
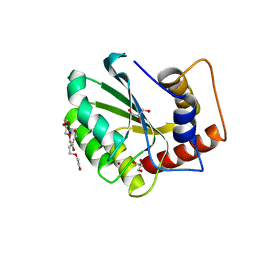 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, GLYCEROL, SODIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQH
 
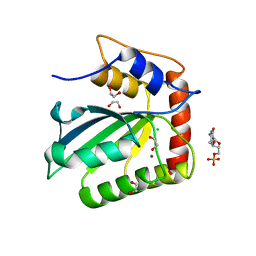 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5U0M
 
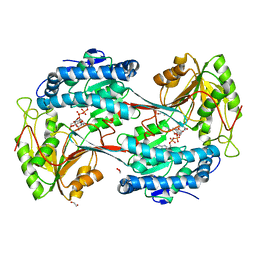 | | Fatty aldehyde dehydrogenase from Marinobacter aquaeolei VT8 and cofactor complex | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, N-succinylglutamate 5-semialdehyde dehydrogenase, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
5CQD
 
 | |
5W8X
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations-Bound to UDP | | Descriptor: | Lipid-A-disaccharide synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
5WLY
 
 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
5WFY
 
 | |
5SWW
 
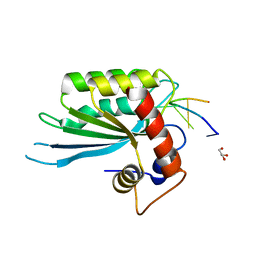 | | Crystal Structure of Human APOBEC3A complexed with ssDNA | | Descriptor: | DNA 15-Mer, DNA dC->dU-editing enzyme APOBEC-3A, GLYCEROL, ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W8N
 
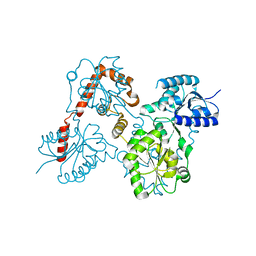 | |
5CQI
 
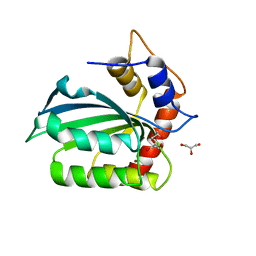 | |
5W8S
 
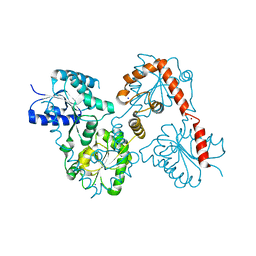 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | Descriptor: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
6E8C
 
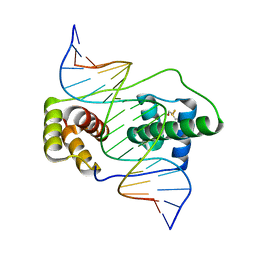 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
5TD5
 
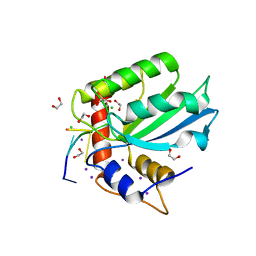 | | Crystal Structure of Human APOBEC3B variant complexed with ssDNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(P*TP*TP*CP*AP*T)-3'), ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5SXH
 
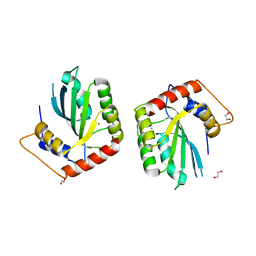 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, ZINC ION | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Sci Rep, 7, 2017
|
|
