1RYA
 
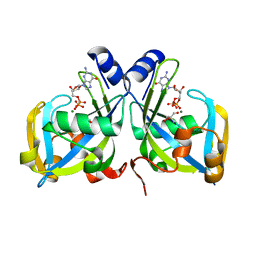 | | Crystal Structure of the E. coli GDP-mannose mannosyl hydrolase in complex with GDP and MG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GDP-mannose mannosyl hydrolase, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Legler, P.M, Mildvan, A.S, Amzel, L.M. | | Deposit date: | 2003-12-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus.
Structure, 12, 2004
|
|
4OVV
 
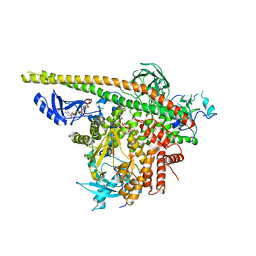 | | Crystal Structure of PI3Kalpha in complex with diC4-PIP2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
2GT4
 
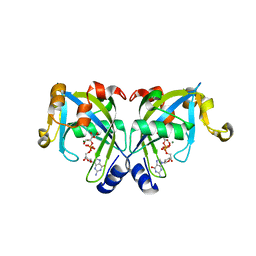 | | Crystal Structure of the Y103F mutant of the GDP-mannose mannosyl hydrolase in complex with GDP-mannose and MG+2 | | Descriptor: | GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, Mildvan, A.S, Amzel, L.A. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
2GT2
 
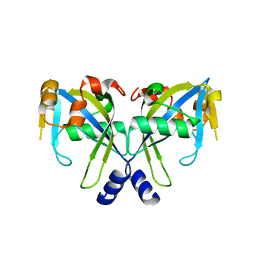 | | Structure of the E. coli GDP-mannose mannosyl hydrolase | | Descriptor: | GDP-mannose mannosyl hydrolase | | Authors: | Gabelli, S.B, Bianchet, M.A, Azurmendi, H.F, MIldvan, A.S, Amzel, L.M. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, NMR, and mutational studies of the catalytic cycle of the GDP-mannose mannosyl hydrolase reaction.
Biochemistry, 45, 2006
|
|
5SW8
 
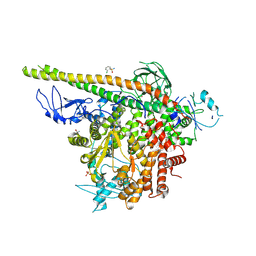 | | Crystal structure of PI3Kalpha in complex with fragments 7 and 11 | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, 2H-indazol-5-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5C7Q
 
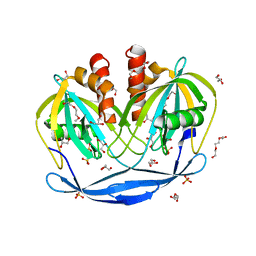 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
5C7T
 
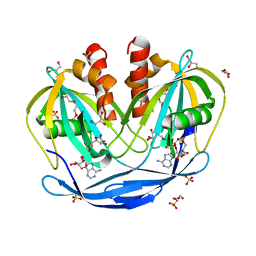 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase in complex with ADP-ribose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5-DIPHOSPHORIBOSE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
4OVU
 
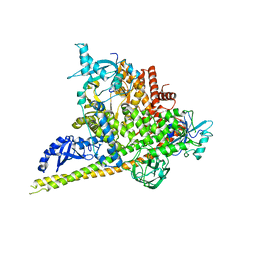 | | Crystal Structure of p110alpha in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
4OVN
 
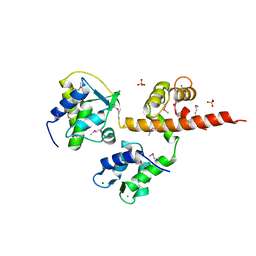 | | Voltage-gated Sodium Channel 1.5 (Nav1.5) C-terminal domain in complex with Calmodulin poised for activation | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Boto, A, Jakoncic, J, Tomaselli, G.F, Amzel, L.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of the NaV1.5 cytoplasmic domain by calmodulin.
Nat Commun, 5, 2014
|
|
4HXX
 
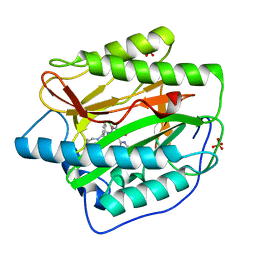 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
4IU6
 
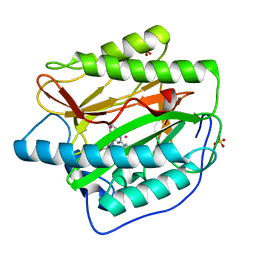 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | Descriptor: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | Deposit date: | 2013-01-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
1YHL
 
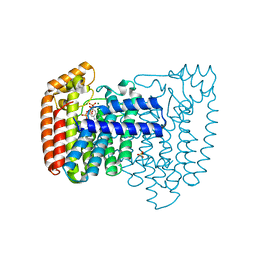 | | Structure of the complex of Trypanosoma cruzi farnesyl diphosphate synthase with risedronate, dmapp and mg+2 | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
1YHK
 
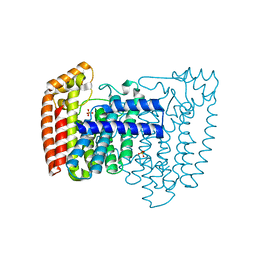 | | Trypanosoma cruzi farnesyl diphosphate synthase | | Descriptor: | SULFATE ION, farnesyl pyrophosphate synthase | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
1YHM
 
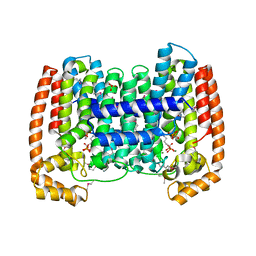 | | Structure of the complex of Trypanosoma cruzi farnesyl disphosphate synthase with alendronate, Isopentenyl diphosphate and mg+2 | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
2M1M
 
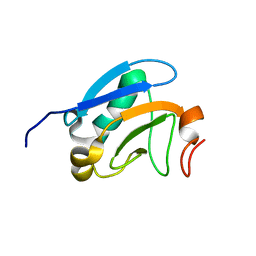 | | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response | | Descriptor: | Auxin-induced protein IAA4 | | Authors: | Kovermann, M, Dinesh, D.C, Gopalswamy, M, Abel, S, Balbach, J. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6MBA
 
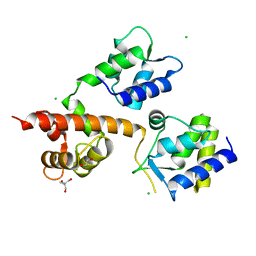 | | Crystal Structure of Human Nav1.4 CTerminal Domain in Complex with apo Calmodulin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONATE ION, ... | | Authors: | Yoder, J, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif.
Nat Commun, 10, 2019
|
|
6MC9
 
 | |
5WJI
 
 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5WJA
 
 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WKW
 
 | | Crystal structure of apo wild type peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WM0
 
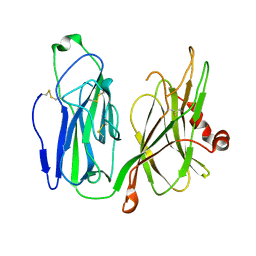 | |
8E0W
 
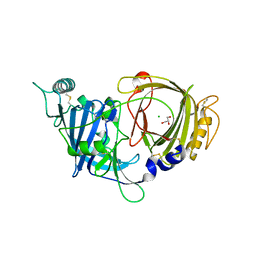 | | Crystal structure of mouse APCDD1 in P1 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E0R
 
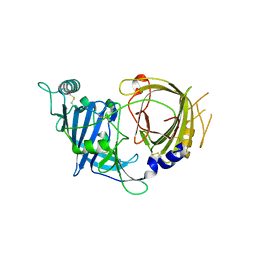 | | Crystal structure of mouse APCDD1 in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein APCDD1 | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2RD0
 
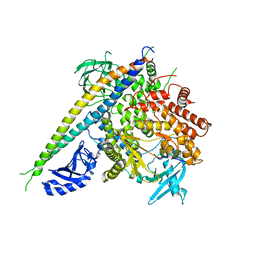 | | Structure of a human p110alpha/p85alpha complex | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations.
Science, 318, 2007
|
|
8FBY
 
 | |
