4QOJ
 
 | | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A | | Descriptor: | FLAVIN MONONUCLEOTIDE, RESVERATROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A
To be Published
|
|
4QOF
 
 | | Crystal structure of fmn quinone reductase 2 AT 1.55A | | Descriptor: | FLAVIN MONONUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of FMN quinone reductase 2 at 1.55A
To be Published
|
|
2FA7
 
 | | Crystal structure of the complex of bovine lactoferrin C-lobe with a pentasaccharide at 2.38 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Singh, N, Jain, R, Jabeen, T, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-12-07 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of the complex of bovine lactoferrin C-lobe with a pentasaccharide at 2.38 A resolution
To be Published
|
|
1BYO
 
 | | WILD-TYPE PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1BYP
 
 | | E43K,D44K DOUBLE MUTANT PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
1ITX
 
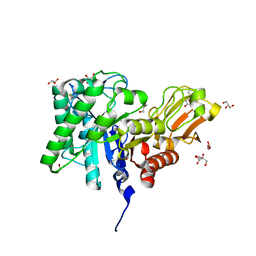 | | Catalytic Domain of Chitinase A1 from Bacillus circulans WL-12 | | Descriptor: | GLYCEROL, Glycosyl Hydrolase | | Authors: | Iwahori, F, Matsumoto, T, Watanabe, T, Nonaka, T. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Three-dimensional structure of the catalytic domain of chitinase A1 from Bacillus circulans WL-12 at a very high resolution
PROC.JPN.ACAD.,SER.B, 75, 1999
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
1WVV
 
 | |
1MF4
 
 | | Structure-based design of potent and selective inhibitors of phospholipase A2: Crystal structure of the complex formed between phosholipase A2 from Naja Naja sagittifera and a designed peptide inhibitor at 1.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2, VAL-ALA-PHE-ARG-SER | | Authors: | Singh, R.K, Vikram, P, Paramsivam, M, Jabeen, T, Sharma, S, Makker, J, Dey, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-08-09 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of specific peptide inhibitors for group I phospholipase A2: structure of a complex formed between phospholipase A2 from Naja naja sagittifera (group I) and a designed peptide inhibitor Val-Ala-Phe-Arg-Ser (VAFRS) at 1.9 A resolution reveals unique features
Biochemistry, 42, 2003
|
|
6THE
 
 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
1WVU
 
 | |
1OYF
 
 | | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol | | Descriptor: | 6-METHYLHEPTAN-1-OL, ACETIC ACID, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-04-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol
To be Published
|
|
3W4K
 
 | | Crystal Structure of human DAAO in complex with coumpound 13 | | Descriptor: | 3-hydroxy-6-(2-phenylethyl)pyridazin-4(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4J
 
 | | Crystal Structure of human DAAO in complex with coumpound 12 | | Descriptor: | 3-hydroxy-5-(2-phenylethyl)pyridin-2(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4I
 
 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
1LN8
 
 | | Crystal Structure of a New Isoform of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Phospholipase A2 | | Authors: | Singh, R.K, Vikram, P, Paramasivam, M, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-05-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a New Form of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution
to be published
|
|
1Q7A
 
 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
1TDV
 
 | | Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, YWAAAA | | Authors: | Singh, N, Jabeen, T, Ethayathulla, A.S, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-specific binding to phospholipase A2:Crystal structure of the complex of PLA2 with a designed peptide Tyr-Trp-Ala-Ala-Ala-Ala at 1.7A resolution
to be published
|
|
1TGM
 
 | | Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution
To be Published
|
|
1T37
 
 | | Design of specific inhibitors of phospholipase A2: Crystal structure of the complex formed between group I phospholipase A2 and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution | | Descriptor: | ACETATE ION, Phospholipase A2 isoform 3, Synthetic peptide | | Authors: | Singh, R.K, Singh, N, Jabeen, T, Makker, J, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-04-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of group I PLA2 with a group II-specific peptide Leu-Ala-Ile-Tyr-Ser (LAIYS) at 2.6 A resolution.
J.Drug Target., 13, 2005
|
|
1OXR
 
 | | Aspirin induces its Anti-inflammatory effects through its specific binding to Phospholipase A2: Crystal structure of the complex formed between Phospholipase A2 and Aspirin at 1.9A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Singh, R.K, Ethayathulla, A.S, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-03 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Aspirin induces its anti-inflammatory effects through its specific binding to phospholipase A2: crystal structure of the complex formed between phospholipase A2 and aspirin at 1.9 angstroms resolution.
J.Drug Target., 13, 2005
|
|
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
6THF
 
 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
