9EOU
 
 | |
9BKK
 
 | | Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Harikumar, K.G, Zhao, P, Cary, B.P, Xu, X, Desai, A.J, Mobbs, J.I, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
9BZB
 
 | | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2
To Be Published
|
|
9B74
 
 | |
9BZN
 
 | | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50 | | Descriptor: | FORMIC ACID, SULFATE ION, class A Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50
To Be Published
|
|
9BHN
 
 | |
8EM7
 
 | | Cryo-EM structure of LRP2 at pH 5.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
9BCZ
 
 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
8YXB
 
 | |
9B75
 
 | |
8EM4
 
 | | Cryo-EM structure of LRP2 at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
7NOS
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 4-bromo-6-(trifluoromethyl)-1H-benzo[d]imidazole. | | Descriptor: | 4-bromanyl-6-(trifluoromethyl)-1~{H}-benzimidazole, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
9B35
 
 | | Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor ionotropic, kainate 2 | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
8WT1
 
 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
8EYO
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 with NADP bound | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, ... | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
9BKJ
 
 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | Descriptor: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
9BHR
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound | | Descriptor: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-{(1E)-3-[(2-methoxyethyl)amino]-3-oxoprop-1-en-1-yl}-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1 | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Al-Awar, R, Krausser, C, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound
To be published
|
|
8YTR
 
 | | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, CopC domain-containing protein, DI(HYDROXYETHYL)ETHER | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Nikolaeva, A.Y, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus
To Be Published
|
|
9ENZ
 
 | |
6XW2
 
 | | Crystal structure of the bright genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein | | Descriptor: | CALCIUM ION, Genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Lazarenko, V.A, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein.
Int J Mol Sci, 21, 2020
|
|
6XWU
 
 | | Crystal structure of drosophila melanogaster CENP-C cumin domain | | Descriptor: | RE68959p | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
7PJE
 
 | | Inhibiting parasite proliferation using a rationally designed anti-tubulin agent | | Descriptor: | Darpin D1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sharma, A, Gaillard, N, Ehrhard, V.A, Steinmetz, M.O. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibiting parasite proliferation using a rationally designed anti-tubulin agent.
Embo Mol Med, 13, 2021
|
|
7NNZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 5-methyl-4-phenylthiazol-2-amine. | | Descriptor: | 5-methyl-4-phenyl-1,3-thiazol-2-amine, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
9EO8
 
 | |
8EYN
 
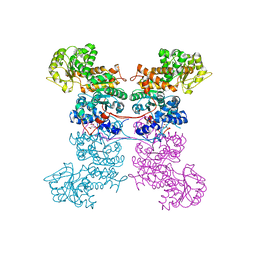 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 in Apo Form | | Descriptor: | CITRIC ACID, NADP-dependent malic enzyme, mitochondrial | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
