4S11
 
 | |
2J2F
 
 | | The T199D Mutant of Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) | | Descriptor: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, FE (III) ION | | Authors: | Guy, J.E, Abreu, I.A, Moche, M, Lindqvist, Y, Whittle, E, Shanklin, J. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Single Mutation in the Castor {Delta}9-18:0- Desaturase Changes Reaction Partitioning from Desaturation to Oxidase Chemistry.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3EY0
 
 | | A new form of DNA-drug interaction in the minor groove of a coiled coil | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, 5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DT)-3', MAGNESIUM ION | | Authors: | Pous, J, Moreno, T, Subirana, J.A, Campos, J.L. | | Deposit date: | 2008-10-17 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Coiled-coil conformation of a pentamidine-DNA complex
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7SVQ
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea in complex with NAD+ | | Descriptor: | L-galactose dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
4W9C
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
3EKV
 
 | |
6PBM
 
 | | Pseudopaline Dehydrogenase with NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline Dehydrogenase | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
5CV4
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/V66H at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Theodoru, A, Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/V66H at cryogenic temperature
To be Published
|
|
2I88
 
 | |
5K92
 
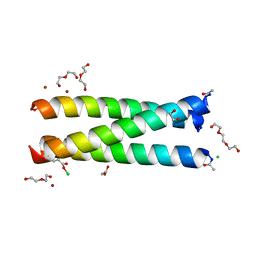 | | Crystal Structure of an apo Tris-thiolate Binding Site in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | Apo-(CSL16C)3, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
3EVE
 
 | |
6PDY
 
 | | Msp1-substrate complex in open conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PEB
 
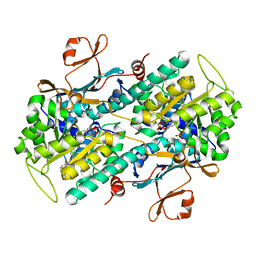 | | Crystal Structure of human NAMPT in complex with NVP-LTM976 | | Descriptor: | N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-3-(pyridin-3-yl)azetidine-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Scaffold Morphing Identifies 3-Pyridyl Azetidine Ureas as Inhibitors of Nicotinamide Phosphoribosyltransferase (NAMPT).
Acs Med.Chem.Lett., 10, 2019
|
|
6I4R
 
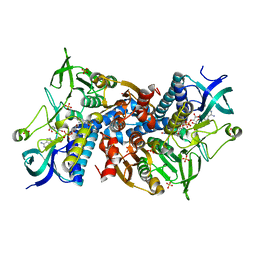 | | Crystal structure of the disease-causing R460G mutant of the human dihydrolipoamide dehydrogenase at 1.44 Angstrom resolution | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Szabo, E, Wilk, P, Bui, D, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
1JXN
 
 | | Crystal Structure of the Lectin I from Ulex europaeus in complex with the methyl glycoside of alpha-L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Audette, G.F, Olson, D.J.H, Ross, A.R.S, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2001-09-07 | | Release date: | 2002-12-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Examination of the Structural Basis for O(H) Blood Group Specificity by Ulex europaeus Lectin I
Can.J.Chem., 80, 2002
|
|
6VRM
 
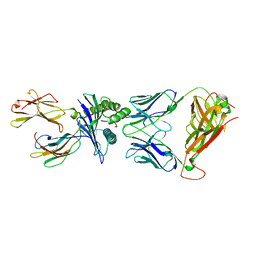 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
7B6R
 
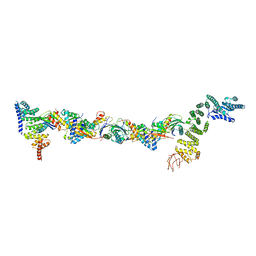 | | Drosophila melanogaster TRAPPIII partial complex: core plus C8 and C11 attached region | | Descriptor: | FI18195p1, GEO08327p1, Probable trafficking protein particle complex subunit 2, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
8A49
 
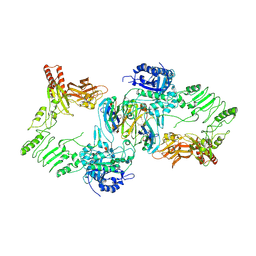 | | Endoglycosidase S in complex with IgG1 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, Secreted endoglycosidase EndoS | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
3E5A
 
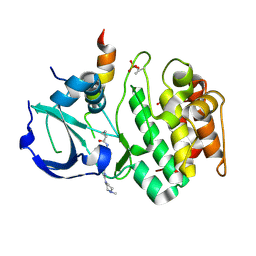 | | Crystal structure of Aurora A in complex with VX-680 and TPX2 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, SULFATE ION, Serine/threonine-protein kinase 6, ... | | Authors: | Zhao, B, Smallwood, A, Lai, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2.
Protein Sci., 17, 2008
|
|
7B6D
 
 | | Drosophila melanogaster TRAPPCore (C1, C2, C2L, C3a/b, C4, C5, C6 subunits) | | Descriptor: | GEO08327p1, Probable trafficking protein particle complex subunit 2, TRAPPC2L, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
2VE3
 
 | | Retinoic acid bound cyanobacterial CYP120A1 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 120, RETINOIC ACID | | Authors: | Kuhnel, K, Ke, N, Sligar, S.G, Schuler, M.A, Schlichting, I. | | Deposit date: | 2007-10-15 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Substrate-Free and Retinoic Acid-Bound Cyanobacterial Cytochrome P450 Cyp120A1.
Biochemistry, 47, 2008
|
|
4IDI
 
 | | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420 | | Descriptor: | GLYCEROL, Oryza sativa Rurm1-related | | Authors: | Wernimont, A.K, Tempel, W, Lew, J, Walker, J, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Bountra, C, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-12 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rurm1-related protein from Plasmodium Yoelii, PY06420
To be Published
|
|
2V8S
 
 | | VTI1B HABC DOMAIN - EPSINR ENTH DOMAIN COMPLEX | | Descriptor: | CLATHRIN INTERACTOR 1, GLYCEROL, VESICLE TRANSPORT THROUGH INTERACTION WITH T-SNARES HOMOLOG 1B | | Authors: | Owen, D.J, McCoy, A.J, Collins, B.M, Miller, S.E. | | Deposit date: | 2007-08-14 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Snare-Adaptor Interaction is a New Mode of Cargo Recognition in Clathrin Coated Vesicles
Nature, 450, 2007
|
|
5DEC
 
 | |
6IC0
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 4 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-pyrimidin-5-yl-pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
