1AMD
 
 | |
5MS1
 
 | | Cowpea mosaic virus top component (CPMV-T) - naturally occurring empty particles | | Descriptor: | Large subunit of Cowpea music virus, Small subunit of cowpea mosaic virus | | Authors: | Hesketh, E.L, Meshcheriakova, Y, Thompson, R.F, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structures of a naturally empty cowpea mosaic virus particle and its genome-containing counterpart by cryo-electron microscopy.
Sci Rep, 7, 2017
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
6S2Z
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Brassica oleracea var. Botrytis with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-Soluble Chlorophyll Protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
5B6W
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 16 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5B6Z
 
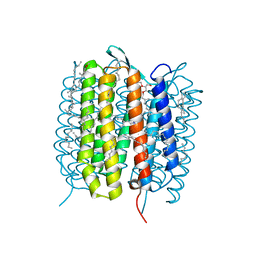 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 1.725 ms us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
8QP5
 
 | | Release Complex: BAM bound EspP (SurA released) | | Descriptor: | Chaperone SurA,Serine protease EspP, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Fenn, K.L, Ranson, N.A. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15, 2024
|
|
8WOV
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8QPV
 
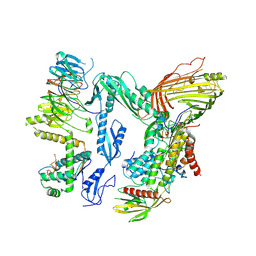 | | Handover Complex: BAM bound OmpX and extended SurA | | Descriptor: | Chaperone SurA,Outer membrane protein X, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Fenn, K.L, Ranson, N.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15, 2024
|
|
3LGI
 
 | |
5MSH
 
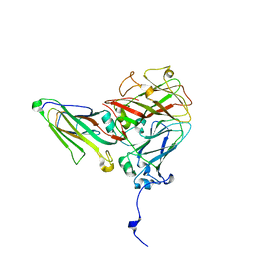 | | Cowpea mosaic virus top component (CPMV-T) - naturally occurring empty particles | | Descriptor: | Cowpea mosaic virus large subunit, Cowpea mosaic virus small subunit | | Authors: | Hesketh, E.L, Meshcheriakova, Y, Thompson, R.F, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structures of a naturally empty cowpea mosaic virus particle and its genome-containing counterpart by cryo-electron microscopy.
Sci Rep, 7, 2017
|
|
1AIF
 
 | | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB FROM MOUSE | | Descriptor: | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (HEAVY CHAIN), ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (LIGHT CHAIN) | | Authors: | Ban, N, Escobar, C, Hasel, K, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-11-14 | | Release date: | 1997-02-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an anti-idiotypic Fab against feline peritonitis virus-neutralizing antibody and a comparison with the complexed Fab.
FASEB J., 9, 1995
|
|
4XKE
 
 | | Crystal structure of hemagglutinin from Taiwan (2013) H6N1 influenza virus in complex with 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Tzarum, N, Zhu, X, Wilson, I.A. | | Deposit date: | 2015-01-11 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structure and Receptor Binding of the Hemagglutinin from a Human H6N1 Influenza Virus.
Cell Host Microbe, 17, 2015
|
|
6S4O
 
 | | The crystal structure of glycogen phosphorylase in complex with 9 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
4EIY
 
 | | Crystal structure of the chimeric protein of A2aAR-BRIL in complex with ZM241385 at 1.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Liu, W, Chun, E, Thompson, A.A, Chubukov, P, Xu, F, Katritch, V, Han, G.W, Heitman, L.H, Ijzerman, A.P, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for allosteric regulation of GPCRs by sodium ions.
Science, 337, 2012
|
|
7K34
 
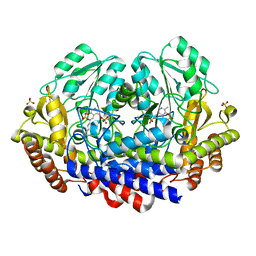 | |
5MAE
 
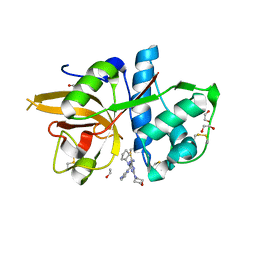 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-car boxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, [4-[cyclopentyl(pyrazin-2-ylmethyl)amino]-6-morpholin-4-yl-1,3,5-triazin-2-yl]methylideneazanide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
5FFL
 
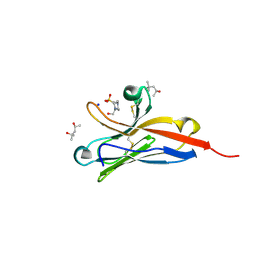 | |
7JMF
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 42 - State 6 (S6) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7QYO
 
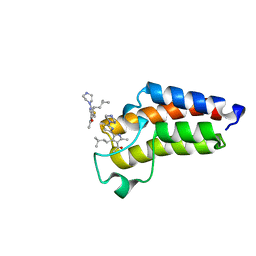 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 79 | | Descriptor: | 1-[2-methyl-4-(3-methylbutyl)-5-(2-piperazin-1-yl-1,3-thiazol-4-yl)-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-28 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.983 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
6ECL
 
 | | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[4-methoxy-3-[[5-methyl-4-(phenylmethyl)-1,2,4-triazol-3-yl]sulfanylmethyl]phenyl]ethanone, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase
To be published
|
|
8G8I
 
 | |
5FGB
 
 | | Three dimensional structure of broadly neutralizing human anti - Hepatitis C virus (HCV) glycoprotein E2 Fab fragment HC33.4 | | Descriptor: | Anti-HCV E2 Fab HC84-1 heavy chain, Anti-HCV E2 Fab HC84-1 light chain, GLYCEROL, ... | | Authors: | Girard-Blanc, C, Rey, F.A, Krey, T. | | Deposit date: | 2015-12-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Antibody Response to Hypervariable Region 1 Interferes with Broadly Neutralizing Antibodies to Hepatitis C Virus.
J.Virol., 90, 2016
|
|
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6S07
 
 | | Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen. | | Descriptor: | Abz-ALA-THR-THR-PRO-LEU-CYS-GLY-PRO-SER-ARG-ALA-SER-ILE-LEU-SER-GLY-ARG, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Leisinger, F, Miarzlou, D.A, Seebeck, F.P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Chem Sci, 10, 2019
|
|
