3FDG
 
 | | The crystal structure of the dipeptidase AC, Metallo peptidase. MEROPS family M19 | | Descriptor: | Dipeptidase AC. Metallo peptidase. MEROPS family M19, MAGNESIUM ION | | Authors: | Zhang, R, Hatzos, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-25 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the dipeptidase AC, Metallo peptidase. MEROPS family M19
To be Published
|
|
6RH4
 
 | | Human Carbonic Anhydrase II in complex with 4-Nitrobenzenesulfonamide. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-nitrobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.948 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
5TID
 
 | | X-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
1SVD
 
 | | The structure of Halothiobacillus neapolitanus RuBisCo | | Descriptor: | GLYCEROL, Ribulose bisphosphate carboxylase small chain, SULFATE ION, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Pashkov, I, Cannon, G, Williams, E, Tran, K, Yeates, T.O. | | Deposit date: | 2004-03-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Halothiobacillus neapolitanus RuBisCo
To be Published
|
|
5TIY
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase/S-oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5HI6
 
 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
6W2A
 
 | | 1.65 A resolution structure of SARS-CoV 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a, [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
8OTT
 
 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | Descriptor: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
7SHJ
 
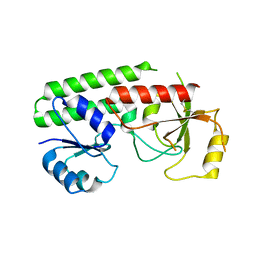 | |
2X38
 
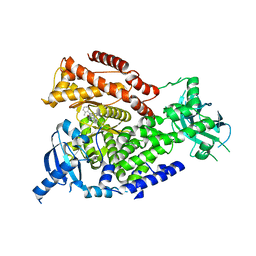 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with IC87114. | | Descriptor: | 2-[(6-AMINO-9H-PURIN-9-YL)METHYL]-5-METHYL-3-(2-METHYLPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
6CBC
 
 | |
4JLU
 
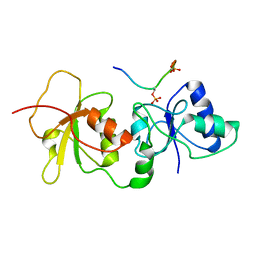 | |
7BHE
 
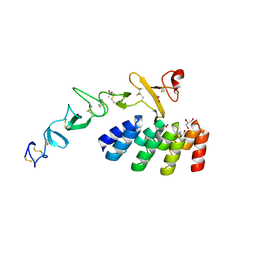 | | DARPin_D5/Her3 domain 4 complex, monoclinic crystals | | Descriptor: | ACETATE ION, DARPin_D5, GLYCEROL, ... | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7BHF
 
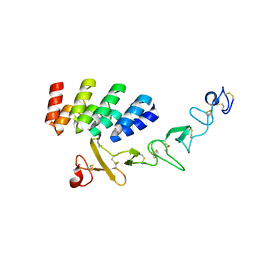 | | DARPin_D5/Her3 domain 4 complex, orthorhombic crystals | | Descriptor: | ACETATE ION, DARPin_D5, Isoform 4 of Receptor tyrosine-protein kinase erbB-3 | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8EZR
 
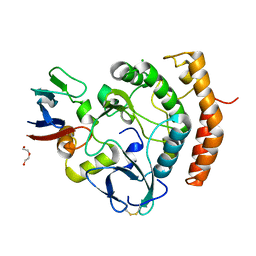 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
4RG6
 
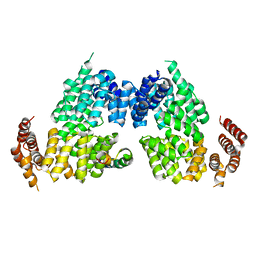 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
9C66
 
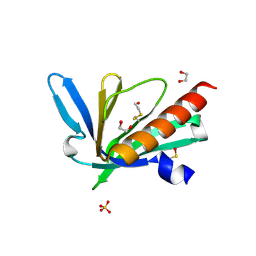 | | Structure of the Mena EVH1 domain bound to the polyproline segment of PTP1B | | Descriptor: | 1,2-ETHANEDIOL, Protein enabled homolog, SULFATE ION, ... | | Authors: | LaComb, L, Fedorov, E, Bonanno, J.B, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the Interaction Landscape of the EVH1 Domain of Mena.
Biochemistry, 63, 2024
|
|
5NNC
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H3K9ac/K14ac) | | Descriptor: | Bromodomain-containing protein 4, Histone H3 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5NIG
 
 | |
2FML
 
 | |
4WNQ
 
 | | THE MOLECULAR BASES OF DELTA/ALPHA-BETA T-CELL MEDIATED ANTIGEN RECOGNITION | | Descriptor: | TCR Variable Beta 2 (TRBV20) chain and TCR constant Beta chain, TCR Variable Delta 1 chain and TCR constant Alpha chain | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
5NNQ
 
 | |
5H8O
 
 | | Crystal structure of an ASC-binding nanobody in complex with the CARD domain of ASC | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, VHH nanobody | | Authors: | Lu, A, Schmidt, F.I, Ruan, J, Tang, C, Wu, H, Ploegh, H.L. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.206 Å) | | Cite: | A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly.
J.Exp.Med., 213, 2016
|
|
7KF5
 
 | |
1T2Y
 
 | | NMR solution structure of the protein part of Cu6-Neurospora crassa MT | | Descriptor: | Metallothionein | | Authors: | Cobine, P.A, McKay, R.T, Zangger, K, Dameron, C.T, Armitage, I.M. | | Deposit date: | 2004-04-23 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Cu metallothionein from the fungus Neurospora crassa
Eur.J.Biochem., 271, 2004
|
|
