4QPR
 
 | |
5CAJ
 
 | | Crystal structure of E. coli YaaA, a member of the DUF328/UPF0246 family | | Descriptor: | BENZAMIDINE, CHLORIDE ION, UPF0246 protein YaaA | | Authors: | Prahlad, J, Lin, J, Wilson, M.A. | | Deposit date: | 2015-06-29 | | Release date: | 2016-02-24 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The DUF328 family member YaaA is a DNA-binding protein with a novel fold.
J.Biol.Chem., 295, 2020
|
|
5K30
 
 | | Crystal structure of methionine gamma-lyase from Citrobacter freundii modified by S-Ethyl-L-cysteine sulfoxide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2016-05-19 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfoxides of sulfur-containing amino acids are suicide substrates of Citrobacter freundii methionine gamma-lyase. Structural bases of the enzyme inactivation.
Biochimie, 168, 2020
|
|
5H2O
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6G28
 
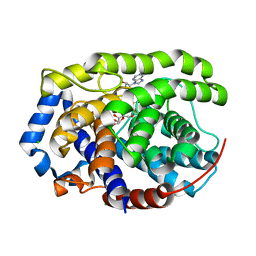 | | Human [protein ADP-ribosylargenine] hydrolase ARH1 in complex with ADP-ribose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, ... | | Authors: | Ariza, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
5MWO
 
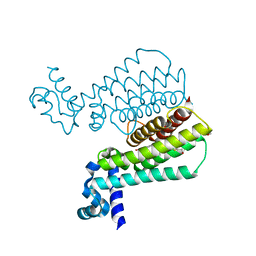 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6G37
 
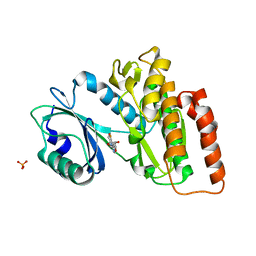 | | Crystal structure of haspin in complex with 5-fluorotubercidin | | Descriptor: | 5-fluorotubercidin, PHOSPHATE ION, Serine/threonine-protein kinase haspin | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5CLB
 
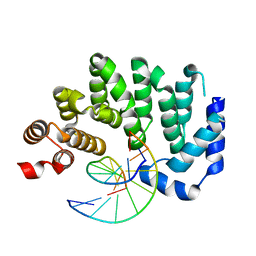 | |
7PL3
 
 | |
5H2L
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 5.25 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
8TIC
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-methylpropyl)-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-19 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
3C92
 
 | | Thermoplasma acidophilum 20S proteasome with a closed gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
5MXZ
 
 | | Kustc0563 Y40F mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cytochrome c-552 Ks_3358, ... | | Authors: | Mohd, A, Barends, T. | | Deposit date: | 2017-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of the c-type cytochrome Kustc0563 from Kuenenia stuttgartiensis
To Be Published
|
|
5CMB
 
 | |
6PBP
 
 | | Pseudopaline Dehydrogenase with (S)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, N-[(1S)-1-carboxy-3-{[(1S)-1-carboxy-2-(1H-imidazol-5-yl)ethyl]amino}propyl]-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
8TJC
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 8a | | Descriptor: | (6M)-1-[(2R)-3,3-dimethylbutan-2-yl]-6-[(5S)-5-methyl-4-oxo-5-phenyl-4,5-dihydro-1H-imidazol-2-yl]-1,3-dihydro-2H-benzimidazol-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-20 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUC
 
 | | Crystal structure of PaIch (Pec1) | | Descriptor: | GLYCEROL, MaoC_dehydrat_N domain-containing protein, NITRATE ION | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural and functional insights of itaconyl-CoA hydratase from Pseudomonas aeruginosa highlight a novel N-terminal hotdog fold.
Febs Lett., 598, 2024
|
|
8TZE
 
 | | Structure of C-terminal half of LRRK2 bound to GZD-824 | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8HPZ
 
 | |
6CBX
 
 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-lysine methyltransferase SMYD2, ... | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497
to be published
|
|
6PBV
 
 | | Crystal structure of Fab668 complex | | Descriptor: | 1,2-ETHANEDIOL, Fab668 heavy chain, Fab668 light chain, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.566 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
5G5V
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-038 | | Descriptor: | (4AS,8AR)-4-(3-{4-[(3R)-3-HYDROXYPYRROLIDINE-1-, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
6PD0
 
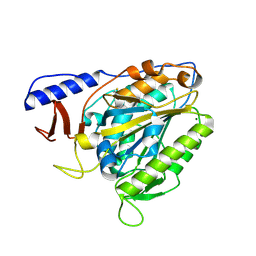 | | Crystal structure of the bacterial cellulose synthase subunit G (BcsG) from Escherichia coli, catalytic domain | | Descriptor: | Cellulose biosynthesis protein BcsG, MAGNESIUM ION, ZINC ION | | Authors: | Anderson, A.C, Brenner, T, Weadge, J.T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | TheEscherichia colicellulose synthase subunit G (BcsG) is a Zn2+-dependent phosphoethanolamine transferase.
J.Biol.Chem., 295, 2020
|
|
8TQO
 
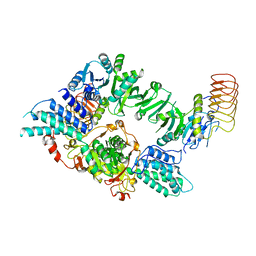 | | Eukaryotic translation initiation factor 2B tetramer | | Descriptor: | Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, Translation initiation factor eIF-2B subunit epsilon, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
8HQ9
 
 | |
