5F0M
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
8Q32
 
 | |
8Q4U
 
 | |
5IEB
 
 | |
6U9E
 
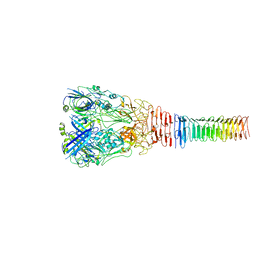 | | Structure of PdpA-VgrG Complex, Lidless | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y, Zhou, Z.H, Horwitz, M.A. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
8Q38
 
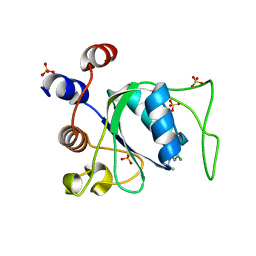 | |
6QXD
 
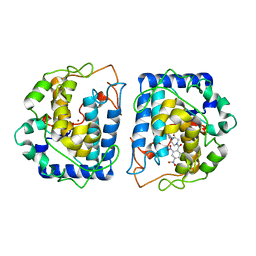 | | Crystal Structure of tyrosinase from Bacillus megaterium with JKB inhibitor in the active site. | | Descriptor: | (2,4-dinitrophenyl)-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]methanone, COPPER (II) ION, Tyrosinase | | Authors: | Deri Zenaty, B, Gitto, R, Pazy, Y, Fishman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Exploiting the 1-(4-fluorobenzyl)piperazine fragment for the development of novel tyrosinase inhibitors as anti-melanogenic agents: Design, synthesis, structural insights and biological profile.
Eur.J.Med.Chem., 178, 2019
|
|
8TA4
 
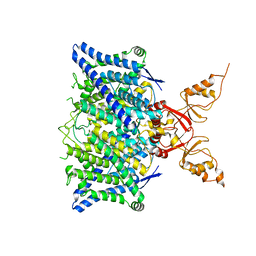 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
5IFA
 
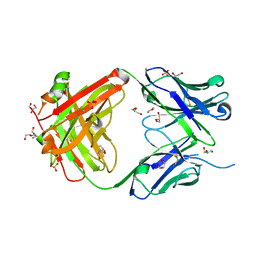 | |
8Q4Q
 
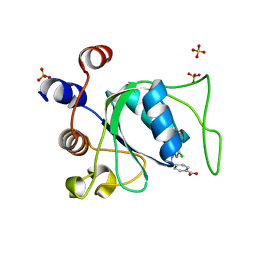 | |
6HNK
 
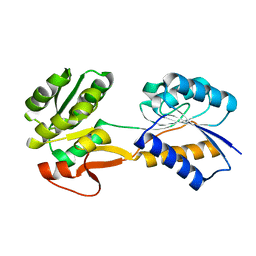 | | The ligand-free, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | ABC-type transport system, sugar-family extracellular solute-binding protein | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
7L97
 
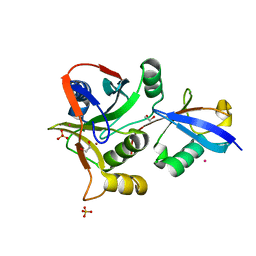 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
7LYW
 
 | |
8Q2T
 
 | |
8Q4R
 
 | |
6HNO
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant S205 - mutant H93A | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bertoletti, N, Heine, A, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mutational and structural studies uncover crucial amino acids determining activity and stability of 17 beta-HSD14.
J.Steroid Biochem.Mol.Biol., 189, 2019
|
|
8Q33
 
 | | Crystal structure of YTHDC1 in complex with Compound 15 (ZA_343) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-[2-[[2-chloranyl-6-(methylamino)purin-9-yl]methyl]phenyl]-2,2,2-tris(fluoranyl)ethanamide | | Authors: | Bedi, R.K, Zalesak, F, Caflisch, A. | | Deposit date: | 2023-08-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
5IGK
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with bromosporine (BSP) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
8Q4V
 
 | | Crystal structure of YTHDC1 in complex with Compound 37 (ZA_356) | | Descriptor: | 2-chloranyl-9-[(3-chlorophenyl)methyl]-~{N}-cyclopropyl-7,8-dihydropurin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Caflisch, A. | | Deposit date: | 2023-08-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
6DGE
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with N5-(1-imino-2-chloroethyl)-L-lysine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~6~-[(1E)-2-chloroethanimidoyl]-L-lysine | | Authors: | Monzingo, A.F, Burstein-Teitelbaum, G, Er, J.A.V, Tuley, A, Fast, W. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Dissection, Optimization, and Structural Analysis of a Covalent Irreversible DDAH1 Inhibitor.
Biochemistry, 57, 2018
|
|
6HSB
 
 | |
6HSU
 
 | |
5WEX
 
 | | Discovery of new selenoureido analogs of 4-(4-fluorophenylureido) benzenesulfonamides as carbonic anhydrase inhibitors | | Descriptor: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Angeli, A, Tanini, D, Peat, T.S, Di Cesare Mannelli, L, Bartolucci, G, Capperucci, A, Ghelardini, C, Supuran, C.T, Carta, F. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
8Q31
 
 | |
6HOK
 
 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
