8W33
 
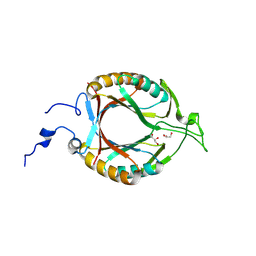 | | Structure of McrD (methyl-coenzyme M reductase operon protein D) from Methanomassiliicoccus luminyensis | | Descriptor: | GLYCEROL, McrD (methyl-coenzyme M reductase operon protein D) | | Authors: | Sutherland-Smith, A.J, Carbone, V, Schofield, L.R, Ronimus, R.S. | | Deposit date: | 2024-02-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of methanogen McrD, a methyl-coenzyme M reductase-associated protein.
Febs Open Bio, 14, 2024
|
|
8V48
 
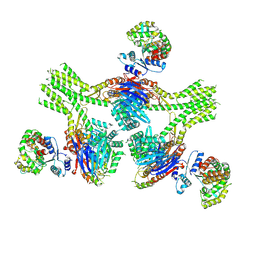 | | CryoEM structure of AriA-AriB complex (Form III) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
7V90
 
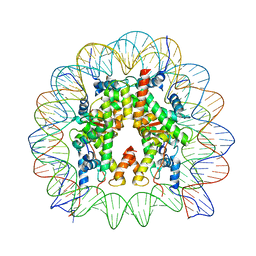 | | Telomeric mononucleosome | | Descriptor: | DNA (145-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
5OXC
 
 | | Structure of Cerulean Fluorescent Protein at 1.02 Angstrom resolution | | Descriptor: | Green fluorescent protein | | Authors: | Gotthard, G, von Stetten, D, Clavel, D, Noirclerc-Savoye, M, Royant, A. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Chromophore Isomer Stabilization Is Critical to the Efficient Fluorescence of Cyan Fluorescent Proteins.
Biochemistry, 56, 2017
|
|
7RPP
 
 | | Crystal structure of human CEACAM1 with GFCC' and ABED face | | Descriptor: | 1,2-ETHANEDIOL, Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of human CEACAM1 oligomerization.
Commun Biol, 5, 2022
|
|
7VA4
 
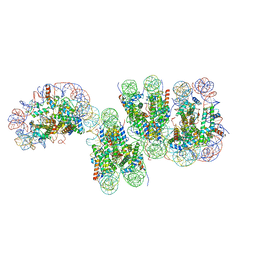 | | Telomeric tetranucleosome in open state | | Descriptor: | DNA (539-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
8OX8
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited "open" conformation | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8BVP
 
 | | Crystal structure of an N-terminal fragment of the effector protein Lpg2504 (SidI) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Restriction endonuclease | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal domain of the effector protein SidI of Legionella pneumophila reveals a glucosyl transferase domain.
Biochem.Biophys.Res.Commun., 661, 2023
|
|
1LAR
 
 | | CRYSTAL STRUCTURE OF THE TANDEM PHOSPHATASE DOMAINS OF RPTP LAR | | Descriptor: | PROTEIN (LAR) | | Authors: | Nam, H.-J, Poy, F, Krueger, N, Saito, H, Frederick, C.A. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the tandem phosphatase domains of RPTP LAR.
Cell(Cambridge,Mass.), 97, 1999
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | Descriptor: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7V9J
 
 | | Telomeric trinucleosome | | Descriptor: | DNA (408-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7WKZ
 
 | | Crystal structure of the HSA complex with mycophenolate and aripiprazole | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, MYCOPHENOLIC ACID, Serum albumin | | Authors: | Kawai, A, Yamasaki, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Structural Basis of the Change in the Interaction Between Mycophenolic Acid and Subdomain IIA of Human Serum Albumin During Renal Failure.
J.Med.Chem., 66, 2023
|
|
6CF2
 
 | | Crystal structure of HIV-1 Rev (residues 1-93)-RNA aptamer complex | | Descriptor: | Anti-Rev Antibody, heavy chain, light chain, ... | | Authors: | Eren, E, Dearborn, A.D, Wingfield, P.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association.
Structure, 26, 2018
|
|
6SWY
 
 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
8C5D
 
 | | Glutathione transferase P1-1 from Mus musculus | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1.
Biomolecules, 13, 2023
|
|
7V9S
 
 | | Telomeric trinucleosome in open state | | Descriptor: | DNA (408-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
5YVW
 
 | |
8OX9
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P active conformation with bound PC | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
3U6I
 
 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 58a | | Descriptor: | Hepatocyte growth factor receptor, N-{3-fluoro-4-[(7-methoxyquinolin-4-yl)oxy]phenyl}-1-[(2R)-2-hydroxypropyl]-5-methyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
7K2V
 
 | |
4Z5C
 
 | | HipB-O3 21mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*CP*GP*TP*AP*GP*AP*GP*CP*GP*GP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
2W51
 
 | | Human mesencephalic astrocyte-derived neurotrophic factor (MANF) | | Descriptor: | PROTEIN ARMET | | Authors: | Parkash, V, Lindholm, P, Peranen, J, Kalkkinen, N, Oksanen, E, Saarma, M, Leppanen, V.M, Goldman, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Conserved Neurotrophic Factors Manf and Cdnf Explains Why They are Bifunctional.
Protein Eng.Des.Sel., 22, 2009
|
|
4Z93
 
 | | BRD4 bromodomain 2 in complex with gamma-carboline-containing compound, number 18. | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-cyclopropyl-5-methyl-1H-pyrazol-4-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-8-methoxy-5H-pyrido[4,3-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2015-04-09 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structure-Based Design of gamma-Carboline Analogues as Potent and Specific BET Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
7LYW
 
 | |
