8QMH
 
 | | Crystal structure of RNA G2C4 repeats in complex with small synthetic molecule ANP77 | | Descriptor: | 3-(7-azanyl-1,8-naphthyridin-2-yl)-2-[(7-azanyl-1,8-naphthyridin-2-yl)methyl]-~{N}-(3-azanylpropyl)propanamide, CHLORIDE ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Kiliszek, A, Ryczek, M, Blaszczyk, L. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
8E8M
 
 | | Mycobacterium tuberculosis RNAP paused elongation complex | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Delbeau, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
5A4B
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | HUMAN ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-06-05 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
8E3N
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5GKG
 
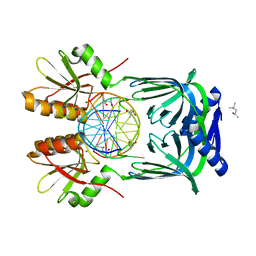 | | Structure of EndoMS-dsDNA1'' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*GP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
8U1C
 
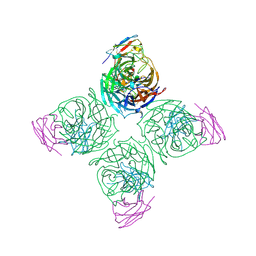 | |
8DVN
 
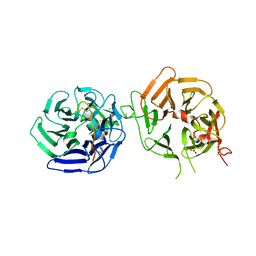 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DSU
 
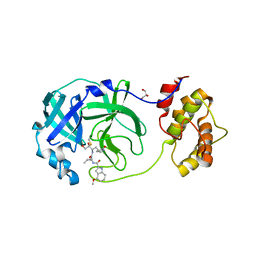 | |
6SAX
 
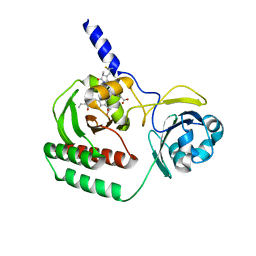 | |
3OPK
 
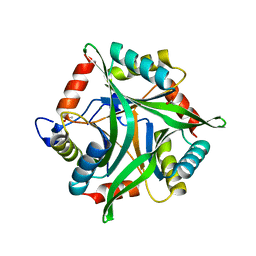 | | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETATE ION, Divalent-cation tolerance protein cutA, MAGNESIUM ION, ... | | Authors: | Nocek, B, Mulligan, R, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
TO BE PUBLISHED
|
|
5I1Y
 
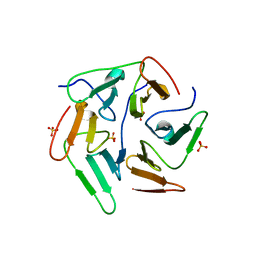 | | NvPizza2-H16S58 with cobalt | | Descriptor: | COBALT (II) ION, SULFATE ION, nvPizz2a-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NvPizza2-H16S58 with cobalt
To Be Published
|
|
6O8M
 
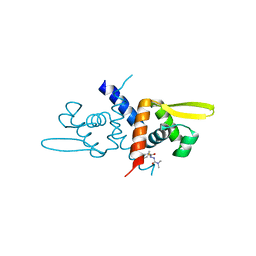 | | Crystal Structure of C9S apo Sulfide-responsive transcriptional repressor (SqrR) from Rhodobacter capsulated bound to diamide (tetramethylazodicarboxamide). | | Descriptor: | N~1~,N~1~,N~2~,N~2~-tetramethylhydrazine-1,2-dicarboxamide, Transcriptional regulator, ArsR family | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for persulfide-sensing specificity in a transcriptional regulator.
Nat.Chem.Biol., 17, 2021
|
|
6M1K
 
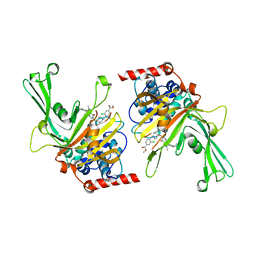 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
8G50
 
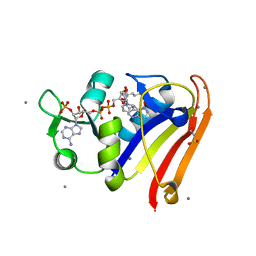 | | E. coli DHFR complex with NADP+ and folate: EF-X excited state model by Laue diffraction (electric field along b axis; 8-fold extrapolation of structure factor differences) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8E7N
 
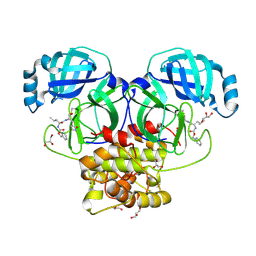 | | Crystal structure of beluga whale Gammacoronavirus SW1 Mpro with GC-376 captured in two conformational states | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, GLYCEROL, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Inhibitor-Bound Main Protease from Delta- and Gamma-Coronaviruses.
Viruses, 15, 2023
|
|
6ODA
 
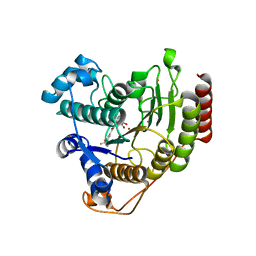 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6OE5
 
 | |
8E7C
 
 | |
4EAQ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate
To be Published
|
|
8QU9
 
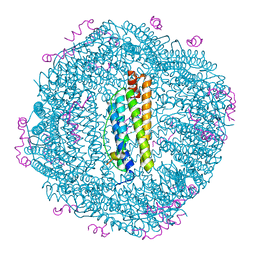 | | Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex | | Descriptor: | FE (III) ION, Ferritin heavy chain, NCOA4 (Nuclear Receptor Coactivator 4) | | Authors: | Hoelzgen, F, Klukin, E, Zalk, R, Shahar, A, Cohen-Schwartz, S, Frank, G.A. | | Deposit date: | 2023-10-15 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for the intracellular regulation of ferritin degradation.
Nat Commun, 15, 2024
|
|
6M6S
 
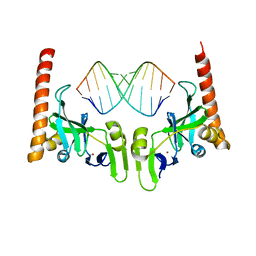 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6VBT
 
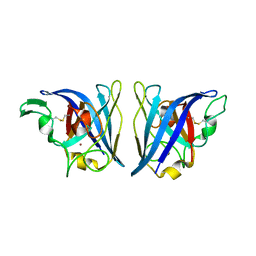 | |
5A38
 
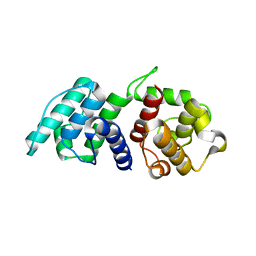 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
8QE8
 
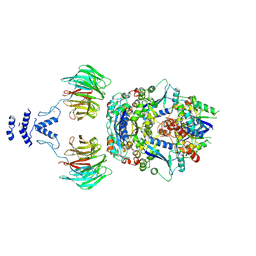 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
5JJ8
 
 | | Crystal Structure of the Beta Carbonic Anhydrase psCA3 isolated from Pseudomonas aeruginosa - alternate crystal packing form | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Pinard, M.A, Kurian, J.J, Aggarwal, M, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Cryoannealing-induced space-group transition of crystals of the carbonic anhydrase psCA3.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
