6QIT
 
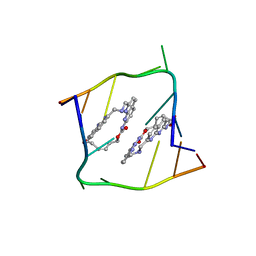 | | Crystal structure of CAG repeats with synthetic CMBL3b compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6X4C
 
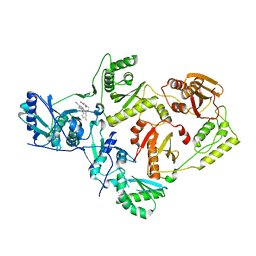 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-5,8-dimethyl-2-naphthonitrile (JLJ658), a Non-nucleoside Inhibitor | | Descriptor: | 7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-5,8-dimethylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6Z7P
 
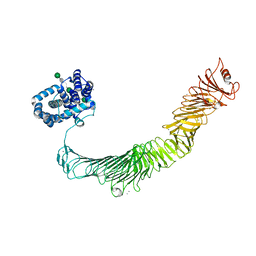 | | Composite model of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | Bharat, T.A.M, von Kugelgen, A. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
7QY2
 
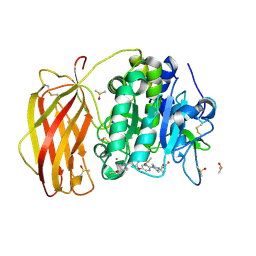 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 2 | | Descriptor: | (2R)-4-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyrimidin-2-yl]piperazin-1-ium-1-yl]-2-methyl-butanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7Q62
 
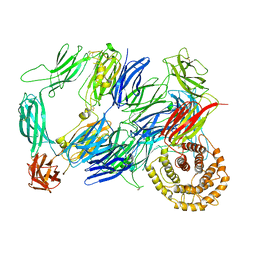 | |
6QDF
 
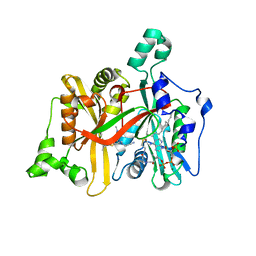 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000096 | | Descriptor: | 3-[[6-tert-butyl-2-[methyl-[(3S)-1-methylpyrrolidin-3-yl]amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QLF
 
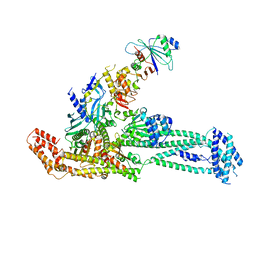 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6WZ0
 
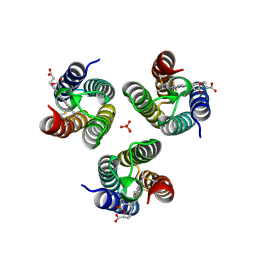 | |
7QY1
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyridin-2-yl]piperazin-1-ium-1-yl]propanoate, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
6QGA
 
 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
7BS1
 
 | | Bovine Pancreatic Trypsin with benzamidine (Room Temperature) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
5NPF
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with beta Cyclophellitol Cyclosulfate probe ME594 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucosylceramidase, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
6Q4U
 
 | | KlenTaq DNA pol in a closed ternary complex with 7-deaza-7-(2-(2-hydroxyethoxy)-N-(prop-2-yn-1-yl)acetamide)-2-dATP | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7QY0
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
8EFZ
 
 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
6YVE
 
 | | Glycogen phosphorylase b in complex with pelargonidin 3-O-beta-D-glucoside | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Drakou, C.E, Gardeli, C, Tsialtas, I, Alexopoulos, S, Mallouchos, A, Koulas, S, Tsagkarakou, A, Asimakopoulos, D, Leonidas, D.D, Psarra, A.M, Skamnaki, V.T. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Crystallography Reveals Binding of Pomegranate Juice Anthocyanins at the Inhibitor Site of Glycogen Phosphorylase: The Contribution of a Sugar Moiety to Potency and Its Implications to the Binding Mode.
J.Agric.Food Chem., 68, 2020
|
|
6YX0
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[2-(4-oxidanylidenebutanoyl)hydrazinyl]methyl]benzoic acid, PWQ-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
5NHY
 
 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
7NTH
 
 | | Structure of TAK1 in complex with compound 54 | | Descriptor: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5NJH
 
 | | Triazolopyrimidines stabilize microtubules by binding to the vinca inhibitor site of tubulin | | Descriptor: | 5-chloranyl-7-[(1~{R},5~{S})-3-methoxy-8-azabicyclo[3.2.1]octan-8-yl]-6-[2,4,6-tris(fluoranyl)phenyl]-[1,2,4]triazolo[1,5-a]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, A, Calvo, G.S, Prota, A.E, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Triazolopyrimidines Are Microtubule-Stabilizing Agents that Bind the Vinca Inhibitor Site of Tubulin.
Cell Chem Biol, 24, 2017
|
|
6Z54
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2003178 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-Dimethyl-6-(oxan-4-yloxy)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003178
To Be Published
|
|
7NTI
 
 | | Structure of TAK1 in complex with compound 22 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7S1K
 
 | | Cfr-modified Escherichia coli stalled ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Tsai, K, Stojkovic, V, Lee, D.J, Young, I.D, Szal, T, Vazquez-Laslop, N, Mankin, A.S, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6Z5J
 
 | | Arrangement of the matrix protein M1 in influenza A/Hong Kong/1/1968 VLPs (HA,NA,M1,M2) | | Descriptor: | Matrix protein 1 | | Authors: | Peukes, J, Xiong, X, Erlendsson, S, Qu, K, Wan, W, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The native structure of the assembled matrix protein 1 of influenza A virus.
Nature, 587, 2020
|
|
5NMG
 
 | | 868 TCR in complex with HLA A02 presenting SLYFNTIAVL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual Molecular Mechanisms Govern Escape at Immunodominant HLA A2-Restricted HIV Epitope.
Front Immunol, 8, 2017
|
|
