6P93
 
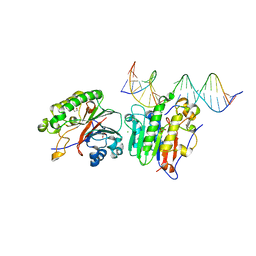 | | Human APE1 K98A AP-endonuclease product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whitaker, A.W, Stark, W.J, Freudenthal, B.D. | | Deposit date: | 2019-06-09 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
Mutagenesis, 35, 2020
|
|
7AHR
 
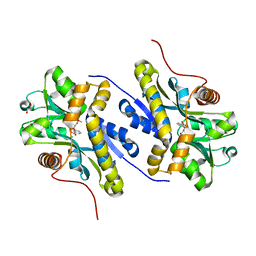 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
1SZE
 
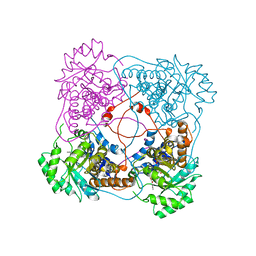 | | L230A mutant flavocytochrome b2 with benzoylformate | | Descriptor: | BENZOYL-FORMIC ACID, Cytochrome b2, mitochondrial, ... | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
5KDO
 
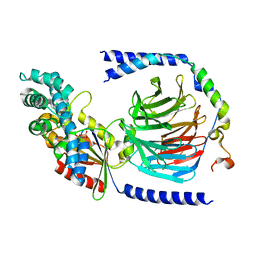 | | Heterotrimeric complex of the 4 alanine insertion variant of the Gi alpha1 subunit and the Gbeta1-Ggamma1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Conserved Hydrophobic Core in G alpha i1 Regulates G Protein Activation and Release from Activated Receptor.
J.Biol.Chem., 291, 2016
|
|
3E2C
 
 | | Escherichia coli Bacterioferritin Mutant E128R/E135R | | Descriptor: | 1,2-ETHANEDIOL, Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wong, S.G, Tom-Yew, S.A.L, Lewin, A, Le Brun, N.E, Moore, G.R, Murphy, M.E.P, Mauk, A.G. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Studies of a Stabilized Subunit Dimer Variant of Escherichia coli Bacterioferritin Identify Residues Required for Core Formation.
J.Biol.Chem., 284, 2009
|
|
5JOZ
 
 | | Bacteroides ovatus Xyloglucan PUL GH43B | | Descriptor: | CALCIUM ION, Non-reducing end alpha-L-arabinofuranosidase BoGH43B | | Authors: | Hemsworth, G.R, Thompson, A.J, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
1T9D
 
 | | Crystal Structure Of Yeast Acetohydroxyacid Synthase In Complex With A Sulfonylurea Herbicide, Metsulfuron methyl | | Descriptor: | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE, Acetolactate synthase, mitochondrial, ... | | Authors: | McCourt, J.A, Pang, S.S, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2004-05-16 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase.
Biochemistry, 44, 2005
|
|
1JGX
 
 | | Photosynthetic Reaction Center Mutant With Thr M 21 Replaced With Asp | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8A3X
 
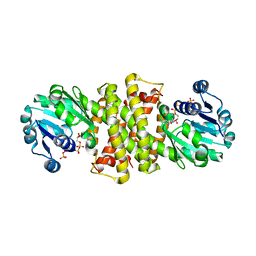 | | Imine Reductase from Ensifer adhaerens in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NAD_binding_2 domain-containing protein, POTASSIUM ION | | Authors: | Gilio, A.K, Grogan, G. | | Deposit date: | 2022-06-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Reductive Aminase Switches to Imine Reductase Mode for a Bulky Amine Substrate.
Acs Catalysis, 13, 2023
|
|
8A82
 
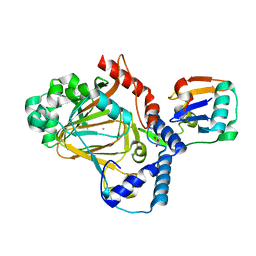 | | Fe(II)/aKG-dependent halogenase OocPQ | | Descriptor: | Cupin_8 domain-containing protein, FE (III) ION, GLYCEROL, ... | | Authors: | Fraley, A.E, Meoded, R.A, Schmalhofer, M, Bergande, C, Groll, M, Piel, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Heterocomplex structure of a polyketide synthase component involved in modular backbone halogenation.
Structure, 31, 2023
|
|
2I59
 
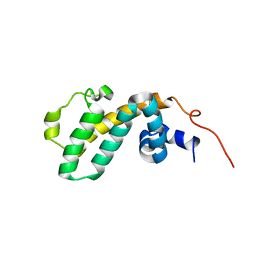 | | Solution structure of RGS10 | | Descriptor: | Regulator of G-protein signaling 10 | | Authors: | Fedorov, O, Higman, V.A, Diehl, A, Leidert, M, Lemak, A, Schmieder, P, Oschkinat, H, Elkins, J, Soundarajan, M, Doyle, D.A, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7T3D
 
 | | CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 222-1C06 mAb heavy chain, ... | | Authors: | Han, J, Ward, A.B. | | Deposit date: | 2021-12-07 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
Nature, 602, 2022
|
|
2W0I
 
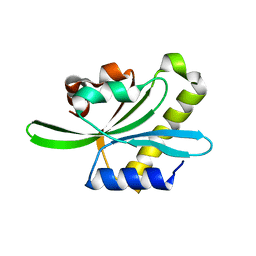 | | Structure Of C-Terminal Actin Depolymerizing Factor Homology (Adf-H) Domain Of Human Twinfilin-2 | | Descriptor: | TWINFILIN-2 | | Authors: | Elkins, J.M, Pike, A.C.W, Salah, E, Savitsky, P, Murray, J.W, Roos, A, von Delft, F, Edwards, A, Arrowsmith, C.H, Wikstrom, M, Bountra, C, Knapp, S. | | Deposit date: | 2008-08-18 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of C-Terminal Actin Depolymerizing Factor Homology (Adf-H) Domain of Human Twinfilin- 2
To be Published
|
|
2VOW
 
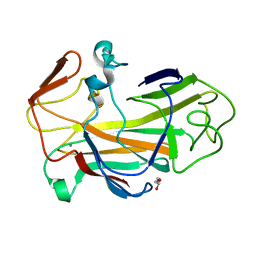 | | An oxidized tryptophan facilitates copper-binding in Methylococcus capsulatus secreted protein MopE. The structure of recombinant MopE to 1.65AA | | Descriptor: | CALCIUM ION, GLYCEROL, SURFACE-ASSOCIATED PROTEIN | | Authors: | Helland, R, Fjellbirkeland, A, Karlsen, O.A, Ve, T, Lillehaug, J.R, Jensen, H.B. | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Oxidized Tryptophan Facilitates Copper Binding in Methylococcus Capsulatus-Secreted Protein Mope.
J.Biol.Chem., 283, 2008
|
|
3E8D
 
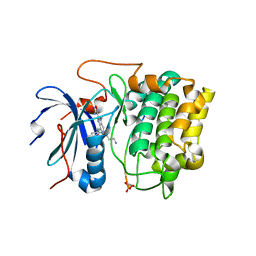 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6PH3
 
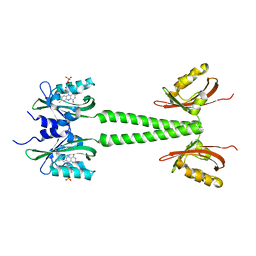 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
8A5Z
 
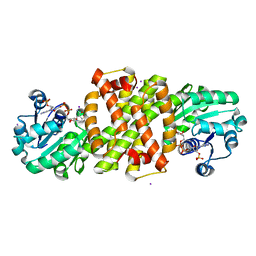 | |
5JZD
 
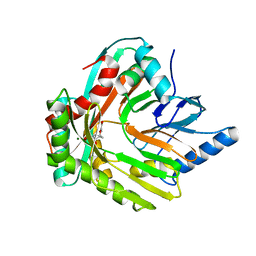 | | A re-refinement of the isochorismate synthase EntC | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, MAGNESIUM ION | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
3WZ2
 
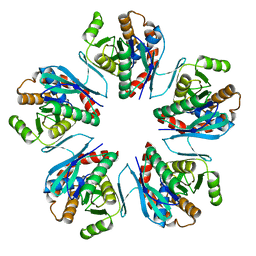 | | Crystal structure of Pyrococcus furiosus PbaA, an archaeal homolog of proteasome-assembly chaperone | | Descriptor: | Uncharacterized protein | | Authors: | Sikdar, A, Satoh, T, Kawasaki, M, Kato, K. | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of archaeal homolog of proteasome-assembly chaperone PbaA
Biochem.Biophys.Res.Commun., 453, 2014
|
|
5DG6
 
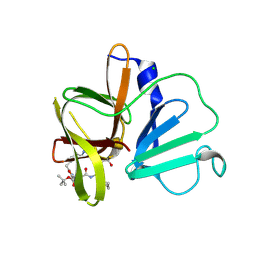 | | 2.35A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (21-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, CHLORIDE ION, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-23-oxa-6,9,14,21,22-pentaazabicyclo[18.2.1]tricosa-1(22),20-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
6PFO
 
 | |
6W5L
 
 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | Descriptor: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
5JYU
 
 | |
5K12
 
 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
1JPW
 
 | | Crystal Structure of a Human Tcf-4 / beta-Catenin Complex | | Descriptor: | BETA-CATENIN, transcription factor 7-like 2 | | Authors: | Poy, F, Lepourcelet, M, Shivdasani, R.A, Eck, M.J. | | Deposit date: | 2001-08-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a human Tcf4-beta-catenin complex.
Nat.Struct.Biol., 8, 2001
|
|
