6UDM
 
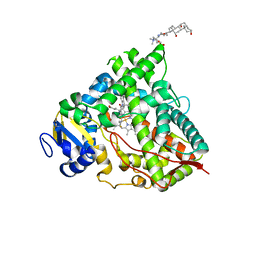 | |
8AZ3
 
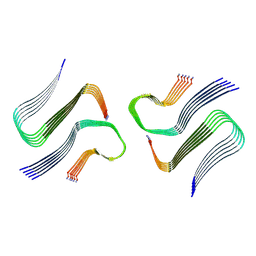 | | IAPP S20G growth-phase fibril polymorph 4PF-CU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
6N1N
 
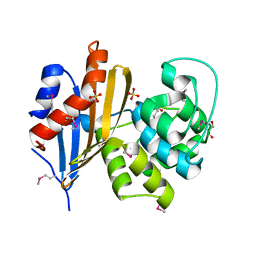 | | Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Michalska, K, Tesar, C, Endres, M, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-09 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386
To Be Published
|
|
8AZ7
 
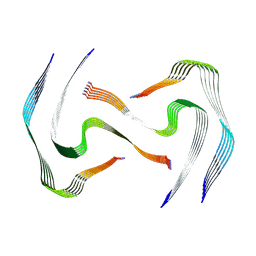 | | IAPP S20G plateau-phase fibril polymorph 4PF-LJ | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
8AZ6
 
 | | IAPP S20G plateau-phase fibril polymorph 4PF-LU | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
7U3B
 
 | | Structure of S. venezuelae GlgX bound to c-di-GMP and acarbose (pH 8.5) | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX, ... | | Authors: | Schumacher, M.A, Tschowri, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
8AZ0
 
 | | IAPP S20G growth-phase fibril polymorph 2PF-L | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Xu, Y, Gallardo, R, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-09-05 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural evolution of fibril polymorphs during amyloid assembly.
Cell, 186, 2023
|
|
6GTU
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with fragment J6 | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, CHLORIDE ION, N-(1,3-benzodioxol-5-ylmethyl)cyclopentanamine, ... | | Authors: | Bertoletti, N, Heine, A, Marchais-Oberwinkler, S, Klebe, G. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Crystallographic Fragment screening and Hit Optimization
To Be Published
|
|
7TVP
 
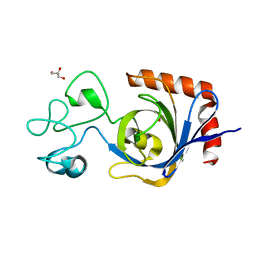 | | Viral AMG chitosanase V-Csn, E157Q mutant, chitotriose complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, GLYCEROL, Viral chitosanase V-Csn E157Q mutant chitotriose complex | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7B8W
 
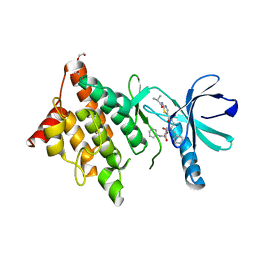 | | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 1 | | Authors: | Lee, H, Yosaatmadja, Y, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Elkins, J.M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470
To Be Published
|
|
7U39
 
 | |
6C6U
 
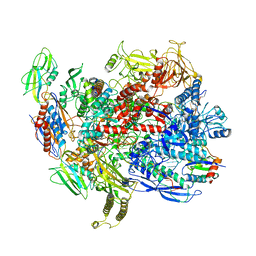 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-DIRECTED RNA POLYMERASE BETA', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6N2P
 
 | | Helical assembly of the CARD9 CARD | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Rohou, A, Arthur, C.P, Dueber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-11-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation.
Nat Commun, 10, 2019
|
|
4XDP
 
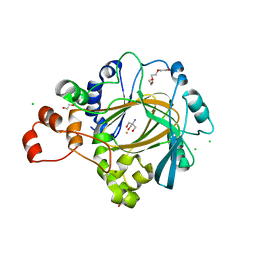 | | Crystal structure of human KDM4C catalytic domain bound to tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
5ZF7
 
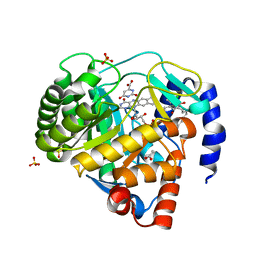 | | Structure of human dihydroorotate dehydrogenase in complex with 277-9-OH | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2E,6E,8S)-8-hydroxy-3,7-dimethylnona-2,6-dien-1-yl]-2-methylbenzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
6HWJ
 
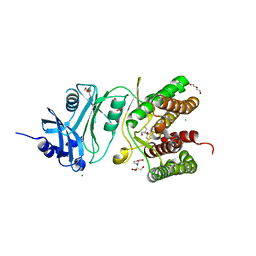 | | Glucosamine kinase (crystal form A) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
Mbio, 10, 2019
|
|
6BOF
 
 | |
5KRI
 
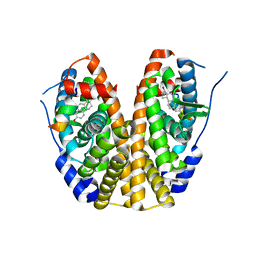 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 16b-benzyl 17b-estradiol | | Descriptor: | (8~{R},9~{S},13~{S},14~{S},16~{R},17~{S})-13-methyl-16-(phenylmethyl)-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthrene-3,17-diol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
7OZ4
 
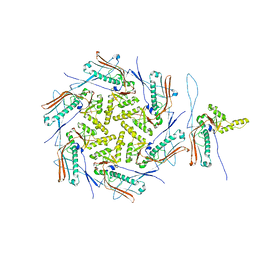 | | Mature capsid of bacteriophage phiRSA1 | | Descriptor: | p2 family phage major capsid protein | | Authors: | Effantin, G, Fujiwara, A, Kawsaki, T, Yamada, T, Schoehn, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High Resolution Structure of the Mature Capsid of Ralstonia solanacearum Bacteriophage phi RSA1 by Cryo-Electron Microscopy.
Int J Mol Sci, 22, 2021
|
|
6C0J
 
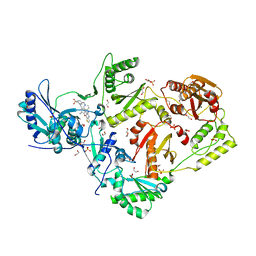 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6N6Q
 
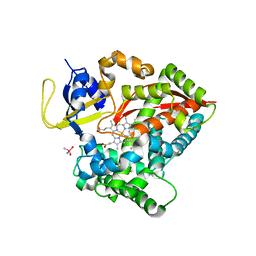 | |
6GIK
 
 | | NMR structure of temporin B L1FK in SDS micelles | | Descriptor: | temporinB_L1FK | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
5HZC
 
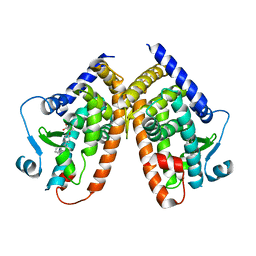 | | Crystal structure of the complex PPARgamma/AL26-29 | | Descriptor: | 2-methyl-2-[4-(naphthalen-1-yl)phenoxy]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A. | | Deposit date: | 2016-02-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode.
Sci Rep, 6, 2016
|
|
6C1K
 
 | | HypoPP mutant with ligand1 | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GUANIDINE, Ion transport protein, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
8V6O
 
 | | Inactivated-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-aminoethyl diphenylborinate, SODIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
