1V4A
 
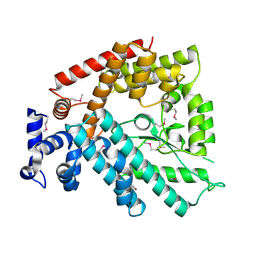 | | Structure of the N-terminal Domain of Escherichia coli Glutamine Synthetase adenylyltransferase | | Descriptor: | Glutamate-ammonia-ligase adenylyltransferase | | Authors: | Xu, Y, Zhang, R, Joachimiak, A, Carr, P.D, Ollis, D.L, Vasudevan, S.G. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Structure, 12, 2004
|
|
7N8V
 
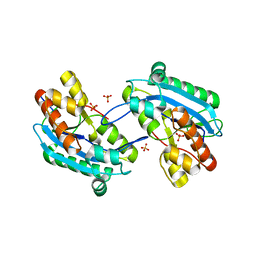 | |
7KJG
 
 | | F96M epi-isozizaene synthase: complex with 3 Mg2+ and BTAC | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
6OXO
 
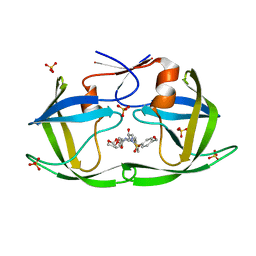 | | HIV-1 Protease NL4-3 WT in Complex with LR2-91 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXP
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
8FRJ
 
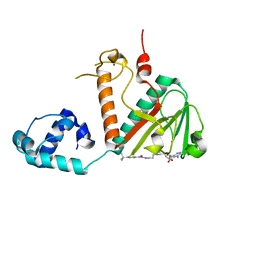 | | Structure of nsp14 N7-MethylTransferase domain fused with TELSAM bound to SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Transcription factor ETV6,Guanine-N7 methyltransferase nsp14 chimera, ZINC ION | | Authors: | Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-01-07 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of SARS-CoV-2 N7-methyltransferase with DOT1L and PRMT7 inhibitors provide a platform for new antivirals.
Plos Pathog., 19, 2023
|
|
6GXX
 
 | | Fab fragment of an antibody selective for alpha-1-antitrypsin in the native conformation | | Descriptor: | FAB 1D9 heavy chain, FAB 1D9 light chain, MAGNESIUM ION | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
7KWX
 
 | | Spermidine N-acetyltransferase SpeG N152L mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7N8W
 
 | |
8Q1C
 
 | | Substrate-free D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-phosphoglucomutase, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
9BZB
 
 | | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2
To Be Published
|
|
1APC
 
 | |
8UTQ
 
 | | KIF1A[1-393] AMP-PNP bound one-head-bound state in complex with a microtubule - class T1L02* | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
5A7F
 
 | | Comparison of the structure and activity of glycosylated and aglycosylated Human Carboxylesterase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LIVER CARBOXYLESTERASE 1, PHOSPHATE ION | | Authors: | Arena de Souza, V, Scott, D.J, Charlton, M, Walsh, M.A, Owen, R.J. | | Deposit date: | 2015-07-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparison of the Structure and Activity of Glycosylated and Aglycosylated Human Carboxylesterase 1.
Plos One, 10, 2015
|
|
6BO3
 
 | | Structure Determination of A223, a turret protein in Sulfolobus turreted icosahedral virus, using an iterative hybrid approach | | Descriptor: | Uncharacterized protein | | Authors: | Sendamarai, A.K, Veesler, D, Fu, C.Y, Marceau, C, Larson, E.T, Johnson, J.E, Lawrence, C.M. | | Deposit date: | 2017-11-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A223, A STRUCTURAL PROTEIN FROM SULFOLOBUS TURRETED ICOSAHEDRAL VIRUS (STIV)
To Be Published
|
|
5JGU
 
 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
9BCZ
 
 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
6TNZ
 
 | | Human polymerase delta-FEN1-PCNA toolbelt | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6R0R
 
 | | Getah virus macro domain in complex with ADPr covalently bond to Cys34 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural polyprotein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S})-2,3,4,5-tetrakis(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6YDJ
 
 | | P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with glucose 6-phosphate and trifluoromagnesate | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
8UTT
 
 | | KIF1A[1-393] P305L mutant AMP-PNP bound two-heads-bound state in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
8Q1F
 
 | | D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with native beta-glucose 1,6-bisphosphate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
6YME
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
