5IOX
 
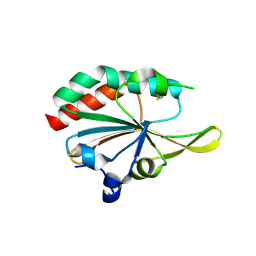 | | Xanthomonas campestris Peroxiredoxin Q - Structure LUss | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
4MJX
 
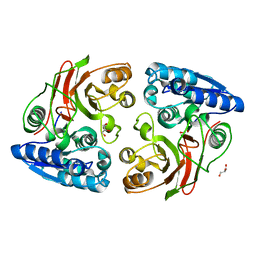 | |
2MP2
 
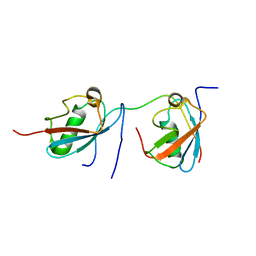 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
7MJS
 
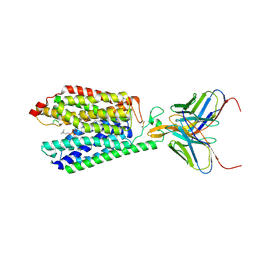 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
8COB
 
 | |
8TDN
 
 | |
5IPT
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 5 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5LZZ
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l (combined) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5A7T
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.4 angstrom resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5LMD
 
 | | The crystal structure of hCA II in complex with a benzoxaborole inhibitor | | Descriptor: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(2-methoxy-5-methyl-phenyl)urea, Carbonic anhydrase 2, ZINC ION | | Authors: | De Simone, G, Alterio, V, Esposito, D, Di Fiore, A. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5F6Q
 
 | | Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames | | Descriptor: | CHLORIDE ION, GLYCEROL, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames
To Be Published
|
|
5ZX8
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Matsumoto, A, Uehara, U, Shimizu, Y, Ueda, T, Uchiumi, T, Ito, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus.
Proteins, 87, 2019
|
|
6H9Y
 
 | | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Outer capsid protein VP4, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
6OY1
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6HAM
 
 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
4QAF
 
 | |
6XHN
 
 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Descriptor: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
7M8T
 
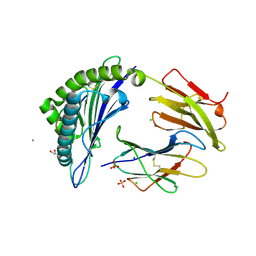 | | Crystal Structure of HLA-A*11:01 in complex with NSASFSTFK, an 9-mer epitope from SARS-CoV-2 spike (S370-378) | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
8A9U
 
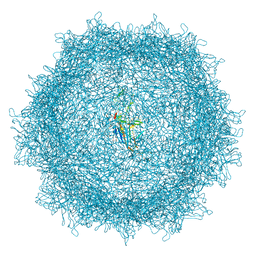 | | Full AAV3B-VP1KO virion | | Descriptor: | Capsid protein VP1 | | Authors: | Arriaga, I, Abrescia, N.G.A. | | Deposit date: | 2022-06-29 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cellular and Structural Characterization of VP1 and VP2 Knockout Mutants of AAV3B Serotype and Implications for AAV Manufacturing.
Hum Gene Ther, 33, 2022
|
|
6RIE
 
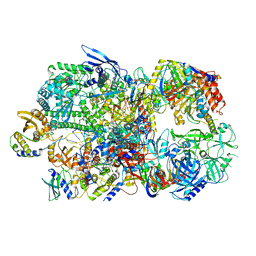 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase co-transcriptional capping complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
5LF1
 
 | | Human 20S proteasome complex with Dihydroeponemycin at 2.0 Angstrom | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
6LFH
 
 | |
5I0R
 
 | | D-cysteine bound C93A mutant of Cysteine Dioxygenase at pH 8 | | Descriptor: | CHLORIDE ION, Cysteine dioxygenase type 1, D-CYSTEINE, ... | | Authors: | Kean, K.M, Driggers, C.M, Karplus, P.A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
6O5G
 
 | | Calmodulin in complex with isomalbrancheamide D | | Descriptor: | (5aS,12aS,13aS)-9-bromo-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7 ,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2019-03-02 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6U66
 
 | | Structure of the trimeric globular domain of Adiponectin | | Descriptor: | Adiponectin, CALCIUM ION, SODIUM ION | | Authors: | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | Deposit date: | 2019-08-29 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
